Using algorithms in healthcare
I want to convince you that our use of machine learning in healthcare, building algorithms that learn for themselves, depends on:
- developing expertise in data analytics and machine learning,
- structuring, generating and aggregating clinically meaningful clinical data, and,
- building robust evaluation processes using a combination of synthetic, randomised controlled and real-life implementation phases.
An algorithm is simply a list of rules to follow in order to solve a problem.
Machine learning
Machine learning uses statistical methods to allow computers to learn from data; in effect, an algorithm is generated by a computer based on data. Modern advances in computationally-intensive methods, such as deep learning, enabled by advances in computing power, have resulted in widespread recent adoption in many domains such as image and speech recognition and excitement about its potential use in healthcare.
Machine learning is already being used in fields outside of image and speech recognition. For example, in 2016, Google DeepMind built a automated recommendation algorithm to improve the energy efficiency of Google’s data centres; the algorithm analyses data from thousands of sensors and is optimised to minimise energy consumption. After two years, with the benefit of real-world experience as well as a range of additional safety measures, the teams took the decision to make the system directly implement its recommendations. As an adaptive algorithm tasked with optimising energy efficiency, the teams have demonstrated an improvement with performance improves over time, as a result of more data being available.
Google DeepMind’s AlphaGo took on and defeated one of the World champions of the game Go. Rather than simply applying a brute force, combinatorial approach to generate the best moves from a finite set of possible moves, the team used deep learning to create a continuously-learning algorithm that improved over time; such learning provided human players with new insights into tactics and strategy as the algorithm used unconventional and unintuitive moves during its play. Their later AlphaGo Zero algorithm, while not needing bootstrapping with even the rules of the game, or from records of previous games, benefitted from a novel approach to reinforcement learning in which it learnt outcomes from games it played against itself, generating data about outcomes in a feedback loop.
But what about machine learning and healthcare?
IBM announced that they would build Watson for Health in 2013, launching services for cancer care that could recommend treatment regimens based on individual patient data and the latest research. During development, IBM acquired a number of companies possessing large amounts of health data, but within four years, the project had been shut down at the M.D. Cancer Centre, in Houston, USA and this year, it was reported that internal IBM reports documented that unsafe and incorrect cancer treatments were being recommended. In writing this blog post, I’ve found conflicting anecdotal reports of its success so it is difficult to conclude anything definitively at present.
Despite such controversies, there have been some successes; Google DeepMind, in a collaboration with Moorfields Eye Hospital, have created an algorithm to interpret optical coherence tomography (OCT) scans, used to examine the structures of the retina in great detail, in order to triage patients by identifying pathology requiring referral. In that paper, the authors compare performance of their algorithm with humans, identifying risks of over- and under-diagnosis.
As such:
- advances in machine learning have created powerful, adaptive, learning algorithms that can outperform humans in niche areas.
- we need need labelled data, including outcome data, in order to teach an algorithm effectively.
- performance of machine learning in healthcare has been subject to hype and relatively few proven success stories
- progress in healthcare, so far, has been associated with taking decisions that have tightly defined scope and that are made in specific clinical contexts.
Algorithms in healthcare
Algorithms are already widely used in medicine, formally and informally.
For example, if you have atrial fibrillation, we can use the CHADS-VASC atrial fibrillation risk score calculator to estimate your risk of developing a stroke and therefore guide preventative treatment; should you be given anticoagulation, or not?
How has such a scoring system been accepted and adopted in healthcare?
This score has been in development over many years, with multiple iterations. Initial calculators such as CHADS2 being published back in 2001 were created by pooling data from 1733 patients with a total of 2121 patient-years of follow-up and have subsequent been refined and validated in different cohorts of patients and adapted as a result of new advances such as the availability of novel anticoagulants that do not need close monitoring. An example of how iterative changes were made in this particular risk algorithm has been documented by Professor Lip from the University of Birmingham, emphasising the importance of ongoing validation and improvement with larger data sets and more representative population samples.
This type of risk score can be generated by examining baseline characteristics and building a statistical model, such as Cox proportional hazards, to identify whether each characteristic has an effect on the outcome measure; such models also tell us the magnitude of that effect. Typically in conventional statistical methodology, the choice of which characteristics to examine is made by the investigators based on existing knowledge about the outcome in question; the approach is hypothesis-driven.
We use the magnitude of effect on outcome to generate a meaningful scoring system which can subsequently be validated as a prediction tool in a particular population. In essence, we predict an endpoint, in this case an outcome, with the presence or absence of characteristics, with information, known at the time of a decision in that population; validation in one population does not mean that an algorithm is appropriate in another. This prediction will itself be uncertain, with the model able to provide prediction of outcome with some degree of confidence; a 95% confidence interval means that we can be 95% certain that our prediction is within the range specified.
Adoption of clinical decision tools therefore result from rigorous process of academic work and ongoing development and validation but is anything more needed?
In fact, adoption of clinical algorithms is also the consequence of a more informal set of processes and cultures that influence whether a tool gains sufficient traction within the wider community. Similarly, even if a tool is widely adopted, the use of the algorithm and any decision made as a result of that algorithm, in any individual clinical scenario, is usually at the discretion of the professional at the time; if it is used then it is used to attempt to reduce the uncertainty about a clinical decision.
As such:
- we already use algorithms
- algorithms become part of clinical practice after going through a set of formal and informal processes, with use dependent on culture, values and perceptions.
- ongoing monitoring and validation is required in order to safely apply an algorithm in a specific population.
- validation of algorithms is currently time-consuming and usually a once-off project, sometimes repeated at intervals. For example, in validating techniques to prevent stroke, there has been dedicated data collection of outcome data, such as whether the patient has had a stroke or not, usually as part of a registry study, because such data is not collected in a meaningful fashion in routine clinical practice.
Heuristics in healthcare
We also use a range of heuristics, sets of informal rules, in our day-to-day clinical work.
When I started as a pre-registration house officer on the neurology ward in 1999, I quickly learned that patients with neuropathy almost certainly have a condition called chronic inflammatory demyelinating polyneuropathy (CIDP), based on my “experience”. Almost every patient I saw with those symptoms had this condition, and my diagnostic sleuthing was calibrated based on this experience. I had quickly and unconsciously developed a heuristic.
Except I was wrong; my calibration was broken because patients with CIDP were the ones who were brought to the ward for their intravenous immunoglobulins and so they were the only patients with neuropathy I saw!
That’s not to say that heuristics are not useful; indeed, heuristics are short-cuts that permit decision making at times of extreme uncertainty, frequently learnt by formal and informal teaching together with experience. For example, a heuristic might be used to take the right action if a life-threatening illness is simply a possibility, even if improbable. In many situations, we chain heuristics together, so that we don’t aim to make a diagnosis in a single cognitive step but instead using a chain of multiple, adaptive heuristics.
For example, I might think you have migraine, but you are 65 years old and your headaches are waking you at night and I proceed to arrange a brain scan. From a purely Bayesian point-of-view, the probability of you having a brain tumour is low; this a priori probability is a consequence of dealing with rarer disorders. My use of red flags, a set of heuristics that, we hope, prompt me to arrange an investigation to reduce the uncertainty in diagnosis in any individual patient, will result in lots of normal scans in the patients that I see. I accept a high rate of negative scans to increase my own sensitivity in identifying a patient with a serious underlying neurological disease; missing such a diagnosis has grave implications for the patient and I endeavour to calibrate my assessment to minimise the possibility, for that specific issue in that specific context.
You might think that heuristics used in clinical practice have been validated in the same way as any other decision aid, but frequently, there is very little data on the positive or negative predictive value of such tools. In addition, humans are subject to a range of cognitive biases (see “Thinking fast and slow” by Prof. Daniel Kahneman) which results in failures of our heuristics. For example, in confirmation bias, we may place greater emphasis on new information if it confirms a pre-existing belief or conclusion.
As such:
- humans use heuristics to help them make decisions, particularly at times of high uncertainty
- humans are prone to a range of biases which result in mistaken decisions
- we will benefit from understanding more about our own decision-making and improving the heuristics we use in daily clinical work; many of our own heuristics would benefit from further evaluation
- specific heuristics are useful, even if of low predictive value, if they increase sensitivity to identify life-threatening illness, in specific clinical contexts.
Becoming data-driven
Decision-making in healthcare, whether by human or machine, whether for making a decision or evaluating a prior decision, needs clinically meaningful data.
What kinds of health data do we have?
- data used for administration, e.g. hospital administrative data
- data for individual clinical care (traditionally paper based), e.g. medical records
- data used for quality improvement and assessment of interventions in real-life environments, e.g. data acquired during specific clinical audit or service improvement projects
- data used for for specific clinical research
- data from the patient, either directly or via their own smart devices
These data are usually separated in silos. It is uncommon for research studies to make use of real-life clinical data and similarly, research data is not usually made available for routine direct care purposes. Similarly, how can we evaluate quality-of-life data without understanding a patient’s long-term health conditions or surgical procedures?
In addition, each type of data is, itself, highly fragmented. For example, data from one research project are held separately to those from another, and even data for direct clinical care usually collected and stored on a per-organisation basis, shared in only a limited fashion, if at all, between organisations providing care for a single patient.
The problems are compounded by the fact that data relating to direct care is frequently paper-based. That’s why most hospitals cannot tell you how many patients with, for example, motor neurone disease , their teams look after or indeed their outcomes, without a project to specifically look at answering those questions for a set period of time.
Even when healthcare professionals use electronic means of capturing information, such as electronic health record software, much of that information is not recorded in a way that makes it easily usable, neither by human nor machine, because it is unstructured or, at best, semi-structured.
For many users, those electronic health record systems are essentially monolithic so that user interface code, business logic and backend data storage is proprietary and must be integrated with other systems to achieve interoperability. When such systems need to be replaced, data must be migrated from the old to the new. We now recognise the benefits of separating data and its structure from the software code that operates upon that data; in healthcare, our data and its structure should be domain-driven given the complex, adaptive environment in which we work and our code can be stateless and often lightweight and ephemeral, particularly those components which are user-facing and most subject to change.
As such:
- we need clinically meaningful data to be recorded and used to support a range of purposes, including: clinical decision making for the care of individual patients, managing our services, quality improvement and clinical research.
- as we capture clinically meaningful data during many different processes and for many different purposes, we must routinely aggregate information of different types, from different sources.
- we need open data standards to build a complete holistic, aggregated view of an individual patient or service; much information is not interpretable without understanding information from elsewhere or from different processes.
- we need semantic interoperability so that we can exchange and combine information from multiple sources.
- we need to move away from procuring ‘full-stack’ applications that combine user interface code, business logic and data storage and move to lightweight, ephemeral user-facing applications each providing different perspectives on the same logical, structured healthcare record.
- as a result, we need to consider the benefit of an open platform, made up of open-source implementations of open standards.
Evaluation and closed feedback loops
We need to consider multiple ways to evaluate our decisions, whether those decisions were made by human or machine.
For human students, we now use simulation at medical schools to help perform and learn in a safe, controlled environment; our human students are exposed to synthetic and real data in order to learn. In my role in assessments for the undergraduate medical course in Cardiff, I helped build a large multiple choice question bank so that we could implement continuous assessment, guiding learning through assessments and feedback on performance at multiple times during the academic year. Our human education has focused on creating a feedback loop in order to help learners improve. Contrived data are useful in testing that a clinician is safe; we might issue a “yellow card” to a student missing a classic presentation of septicaemia in an examination and that student might fail, even if their overall score is in the pass range.
In pharmaceutical drug development, sequential processes are used ranging from drug discovery, preclinical and clinical research, review and post-marketing surveillance. Each step examines the potential or actual efficacy of a drug; initially in models, then in control subjects, then in selected patients and then in real-life clinical environments. Each step makes use of different approaches and provides different insights into the performance of that drug.
In clinical research, such as a randomised controlled trial, we attempt to avoid biases; for example, if we only evaluate an intervention, such as a new drug, in real-life clinical practice, we would fail to control for clinicians inadvertently giving a new treatment only to a most healthy subset of patients. A controlled trial means that we control for spurious differences between a population that receives an intervention and a population that doesn’t, and ensure we remove as many biases as possible in our assessment of that intervention.
The results of randomised controlled trials are usually considered highly generalisable to other populations; however we recognise that this is often not the case when trials exclude patients with, for example, multiple co-morbidities or have failed to recruit patients at the extremes of age.
On the other hand, changes in the way that we deliver care are difficult to evaluate as part of a randomised controlled trial; instead we might adopt an approach of quality improvement. We use these type of methodologies when what we are studying is subject to considerable local variation such as differing local organisational structures; our learning may not necessarily be as generalisable as a randomised controlled trial. Conversely this type of study may provide valuable information about how an intervention performs in the real world.
There are a range of quality improvement methodologies that act to iteratively define, measure, analyse, improve and control; a common feature of all is a cycle of measurement and intervention to examine and improve process and outcome.
Healthcare, as yet, has failed to use technology to transform the way randomised trials and quality improvement are delivered. In modern technology companies, thousands of small randomised trials can be performed every day with closed feedback loops providing rapid assessment of outcome. For example, Google have embedded randomised trials into their software development pipeline, making it possible to run simple trials to assess the effect of changing, for example, font colour, on click-through rates by users.
Routinely collected health data and randomised trials are dependent on a high-quality infrastructure which can collect clinically meaningful data and help to randomise, at any point of clinical equipoise, an individual into a trial of one intervention over another as well as streamlining the process of trial design and ethical approval.
As such:
- underpinning all approaches to evaluation is the collection and analysis of meaningful data, linking eventual outcome to the data known at the time of a decision; in essence, evaluation of any decision is predicated on a closed feedback loop.
- synthetic data is valuable in many circumstances, particularly for beginners.
- randomised controlled trials assess an intervention in a defined group, such as a specific cohort of patients and attempts to control for biases; they may be controlled with a placebo or the best current intervention. Modern trials are frequently complex, expensive and time-consuming and are difficult to run for any complex intervention and usually have very limited follow-up.
- quality improvement approaches assess interventions in real-life contexts but are subject to inherent biases.
- we currently lack a cohesive technical infrastructure that supports the definition, collection and analysis of meaningful, structured clinical data. Likewise, we lack tools to streamline and support the processes needed to undertake randomised trials in humans including design, ethical approval and the day-to-day identification, recruitment, consent and randomisation.
- all clinicians will tell you that we face thousands of situations of clinical equipoise every day, a decision needs to be made in which one choice is no known to be better than another; we must endeavour to build an infrastructure that supports the systematic recruitment of patients into clinical trials as a matter of routine. We are seeing the start of this in oncology.
- such an infrastructure would support our current need for quality improvement and research, but also would support the use of systematic algorithmic decision support in clinical care to shape its development, its evaluation and ongoing post-marketing surveillance of safety.
And so what?
I truly believe that software and data are of vital importance to the future of healthcare.
We need, as professionals and as patients, to have the right information at the right time, in order for us to develop a shared understanding and make the right decisions in any given context. We use data to reduce the uncertainty about our decisions and we need the right tools to create and make sense of those data.
Machine learning is an exciting and continuously-evolving field. I would argue that many of the changes we need to make in order to support the routine development, deployment, evaluation and monitoring of advanced algorithms in healthcare are the same ones that we need to use, right now, for our routine clinical practice. Whether our decisions are made by human or machine, or more likely a combination of both, whether those decisions are for an individual patient and for whole groups of our patients as part of developing our health services, understanding and assessing those decisions by the routine and systematic collection of meaningful data is vital.
We can start to understand what we need to do to achieve this by creating a Wardley map. We can start to build a map by thinking about a value chain, in which we start with user need and try to understand the dependencies:
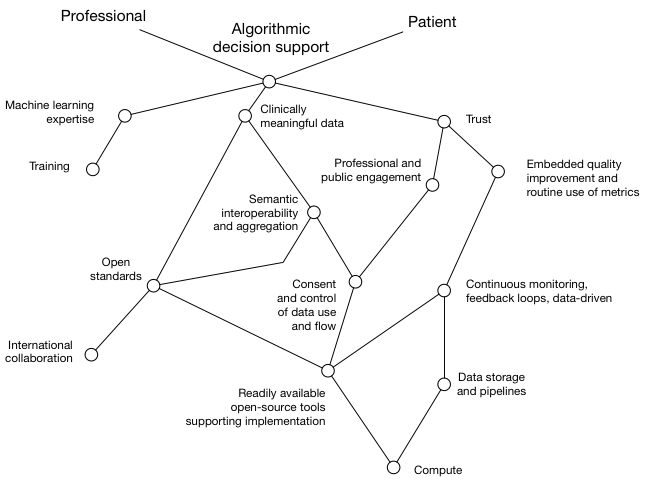
We see that, to provide algorithmic decision support for patients and professionals, we need to consider
- machine learning expertise
- clinically meaningful data
- trust - by the public and professionals.
I have, for each of those, shown what I consider to be the dependencies.
We can plot our value chain on a map, with the horizontal axis giving an indication as to whether that dependency is already available as a commodity, as a product to be bought or rented, as something we are developing or as something that is in its infancy:
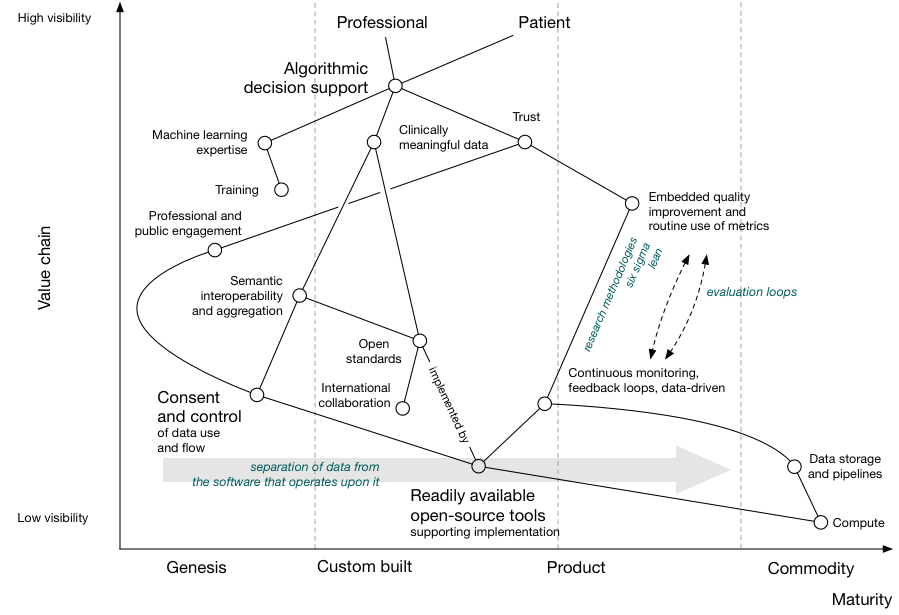
For example, here we see that public and professional trust is dependent on professional and public engagement as well as an evaluation pipeline that creates a continuously monitored feedback loop driven by data. Their location on the map shows that we are only just starting to work on engaging the public on the use of data, while adopting randomised trials and quality improvement such as six sigma in healthcare is more mature but not yet a commodity.
We can add more information. Here, I’ve highlighted whether healthcare organisations or technology companies can provide one of the dependencies:
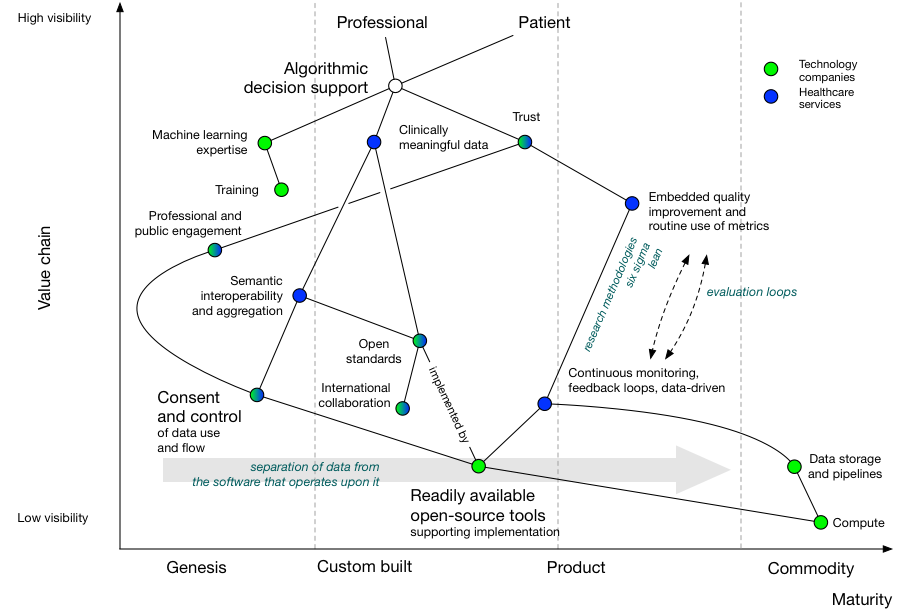
Now we see that machine learning expertise from technology companies must be combined with data expertise from healthcare organisations in order to successfully deliver algorithmic decision support in healthcare.
Finally, we can use our map to work out what we need to do in order to deliver the vision, incorporating an indication of evolution over time:
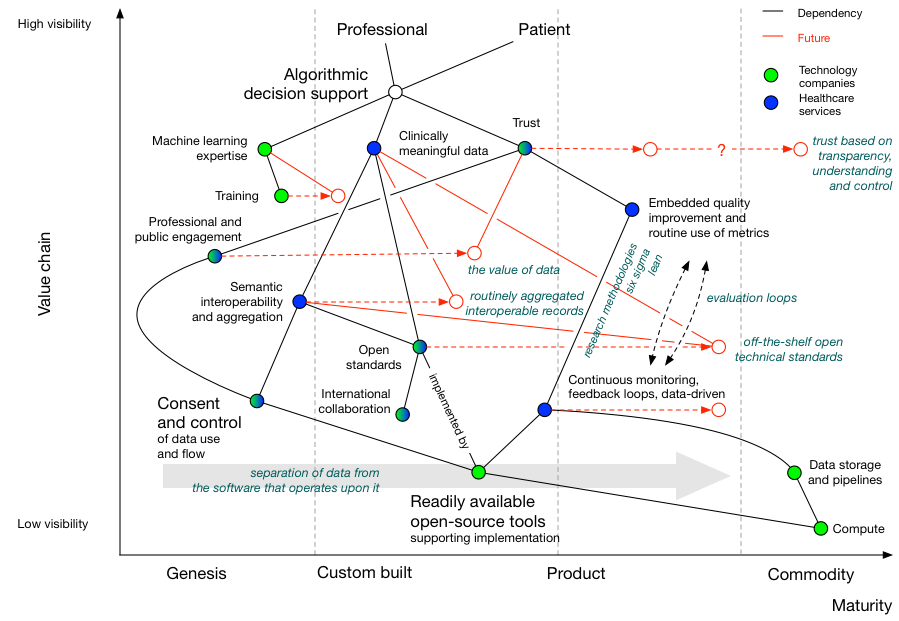
We can conclude from our map that:
- clinically meaningful data is dependent on ongoing work to adopt open standards, and those standards are themselves dependent on an open-source toolchain that makes it easy to implement those open standards.
- aggregation of data from multiple sources is dependent on a scheme of data control and consent, so that citizens can opt-out and opt-in to share data for different purposes. This control and consent, I would argue, is also dependent on readily-available open source solutions.
- continued professional and public engagement will result in an increasing recognition of the value of data
- semantic interoperability, itself dependent on open standards, will result in the creation of routinely aggregated interoperable health records
- trust is dependent on engagement and building an evaluation pipeline supporting development, testing, deployment and real-life evaluation using a variety of processes, themselves supported by an enabling infrastructure
As such, if we want to revolutionise the use of technology in healthcare, we must become data-driven, and to do that, we need to create a suite of, likely open-source, tools that support the implementation of open standards, of consent and of quality improvement/research within day-to-day clinical practice.
Mark
Further reading
I’ve written quite a few posts looking at the benefit of meaningful data in healthcare, the importance of the medical record, diagnostic inference, SNOMED CT and healthcare IT strategy. Here are a few links:
On data:
On creating a next generation cloud health record
- Part one: creative destruction
- Part two: the value of software for healthcare
- Part three: ecosystems and the platform
On platforms:
- Platforms 1/3. Standards and interoperability
- Platforms 2/3. An API-first approach
- Platforms 3/3. What we need to do next
On trust and innovation
- The paradox of ambition
- The paradox of control
- The 5 Os of healthcare IT - objectives, ownership, openness, optimise, organic