So I built an electronic patient record system in my spare time...
This blog post is based on a series of threads I posted on Twitter, which I have refined and expanded for this post.
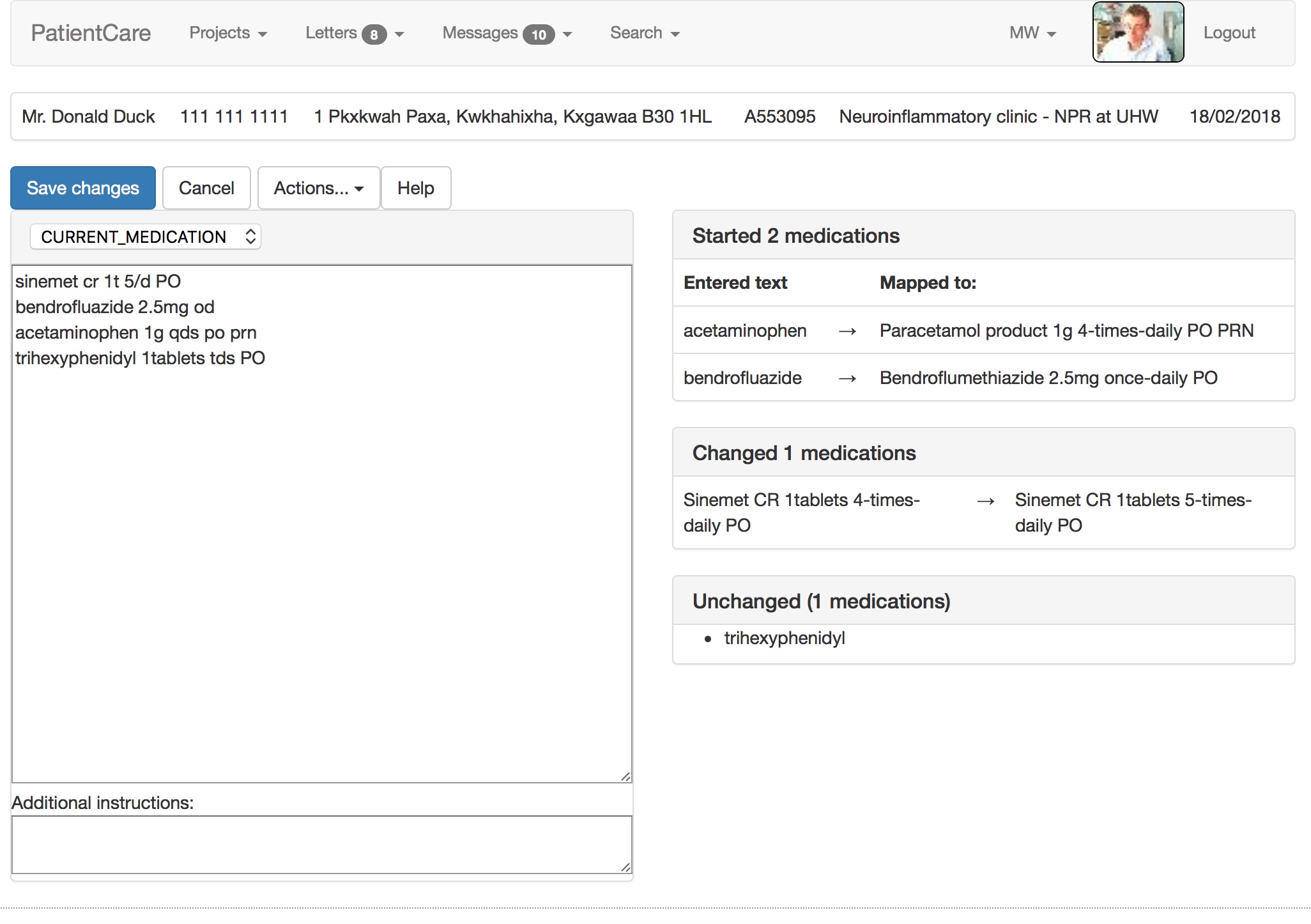
My electronic patient record system uses SNOMED-CT as its lingua-franca. Notice in the video how it maps free text into the UK dm+d (dictionary of medicines and devices), parsing dose route and directions and handling units and equivalence. It suggests a correction when I mistype salbutamol. There is a lot going on under the surface!
Similarly, the entered term acetaminophen is switched to the preferred term, paracetamol. Doctors of a certain age will recognise that bendrofluazide was the term for the drug now known as bendroflumethiazide. Likewise, the doses, frequencies and routes are parsed from free-text into structured, coded, SNOMED-CT data:
Using that information, the software generates a permanent artefact of the consultation, combining human-viewable PDF with structured semantically-useful information for machines.
It calculates in real-time what medications have started, have stopped, are unchanged or have had a change made. If you start doing that, you can track medication changes over a long period of time and show them graphically. The software can calculate the total daily dose, understanding the dose, units and frequencies.
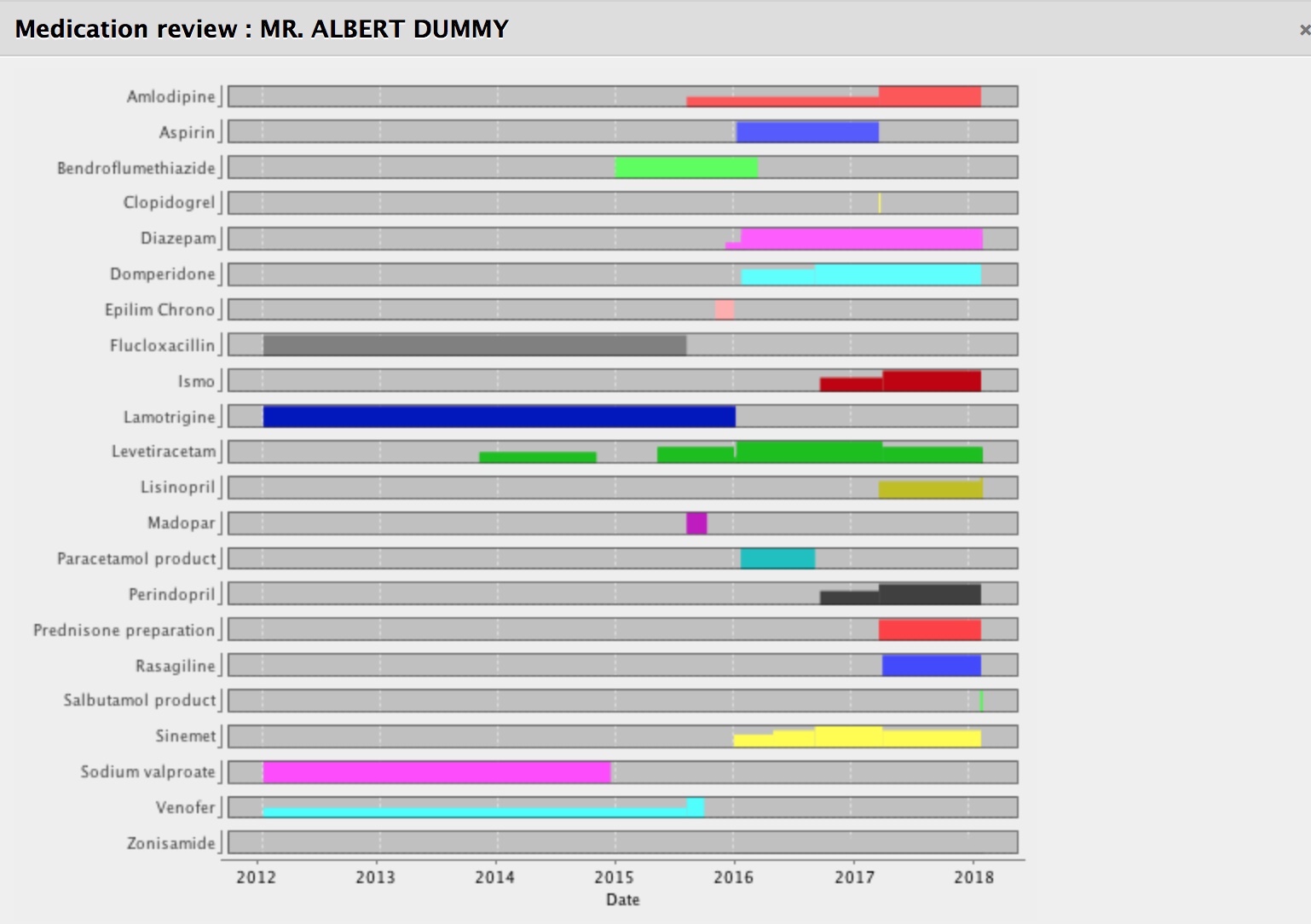
Notice how we can see how the doses have changed or started or stopped over time. For our patients with long-term health conditions, this is very helpful.
Structured documents
I don’t understand how some folk think about structured documents as simply documents with the same types of headings. To me, clinical documents should be structured so that they are useful to humans but also useful to machines. The headings and the information models needed are related but different. We have to make medical data understandable by both humans and machines.
For too long we have created documents from which we hope to derive data. Why not create documents from our data, instead?
So here we record what botulinum toxin injections we have used and generate a letter that goes into the organisation’s document repository…. but we keep the data in a useful and structured format.
Likewise, if I record a patient’s specialist walking assessment then we can use SNOMED-CT as a useful, intermediary structure to allow semantic interoperability. Semantic interoperability means that software written by other people can derive meaning from the data that I exchange with it. Users don’t need to see this, but computers do.
This form, viewable by the professional in the EPR, was populated by the patient entering their information into their iPad.
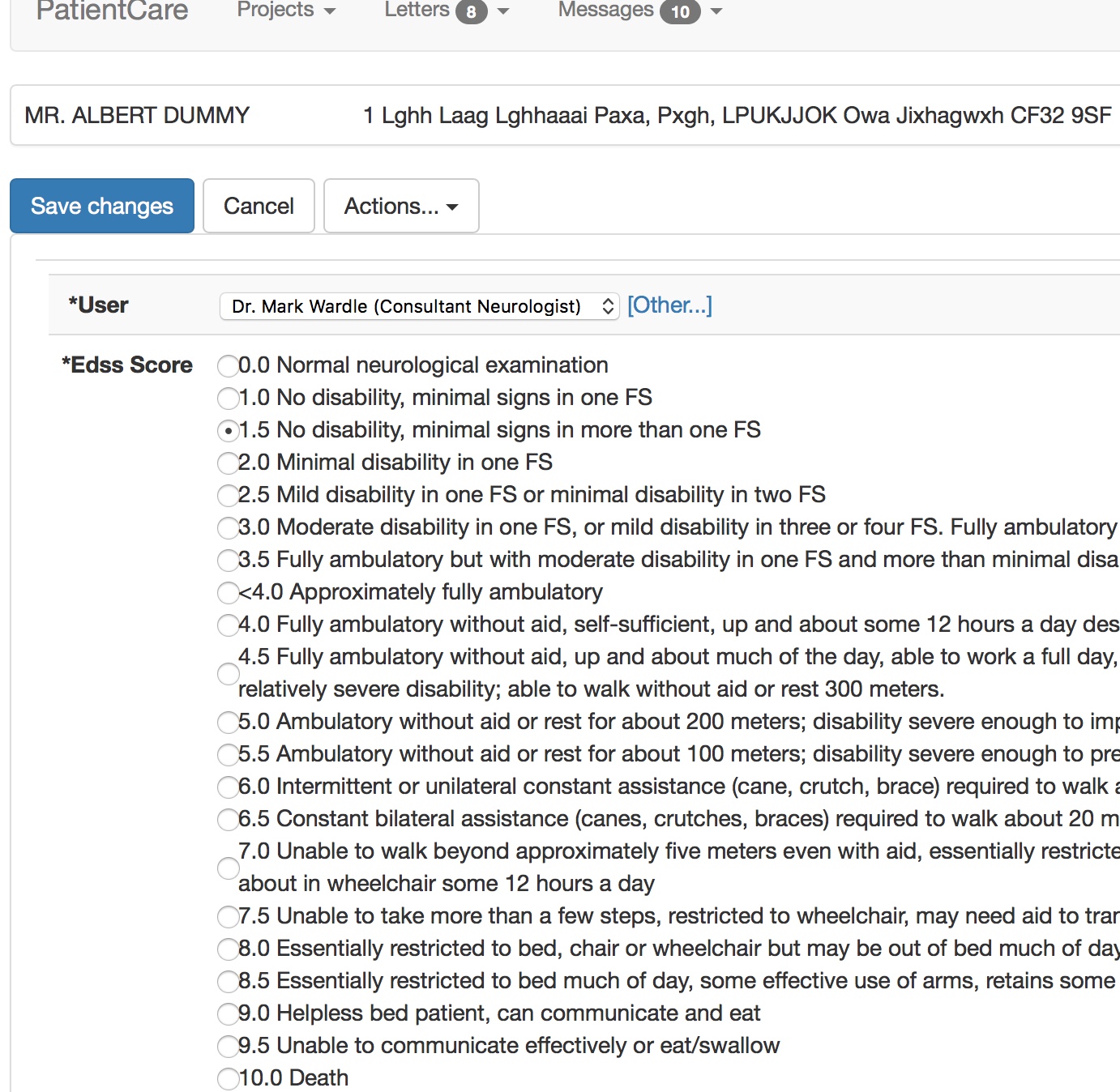
So for interoperability, behind-the-scenes, other software can understand what we’ve done, without necessarily having to understand the Kurtzke EDSS scale. They can, in essence, answer the question “Can this patient walk?” without difficulty.
Here is a simple flat representation of the information from this encounter, recording diagnoses, treatments and outcomes. Even a flat representation allows us to later perform sophisticated searches such as, which patients are currently taking methotrexate. Similarly, I have exposed internal data as HL7 FHIR endpoints using the HAPI libraries.
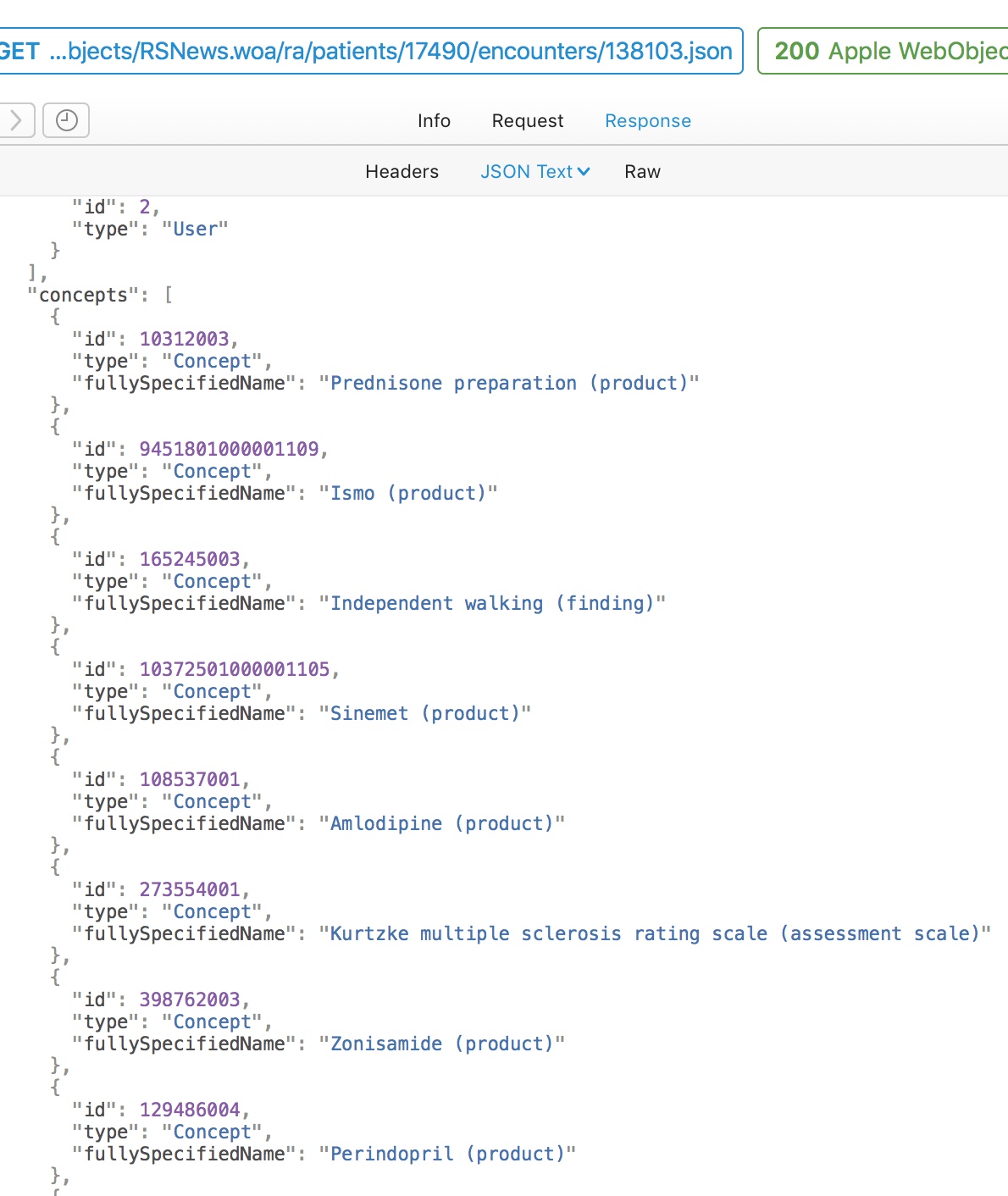
My document therefore consists of a PDF for human consumption and large amounts of structured and semantically useful data that can be processed for that individual or for a whole cohort of patients.
Cohorts
Defining cohorts is important to me, based on service, diagnosis, problem, treatment, intervention or any arbitrary combination thereof. Understanding cohorts is a pre-requisite for understanding outcomes and value. See my previous blog post on the importance of cohorts
Most users have no idea that they are using SNOMED, if you do this right. Here we are entering country of birth, ethnicity and occupation.
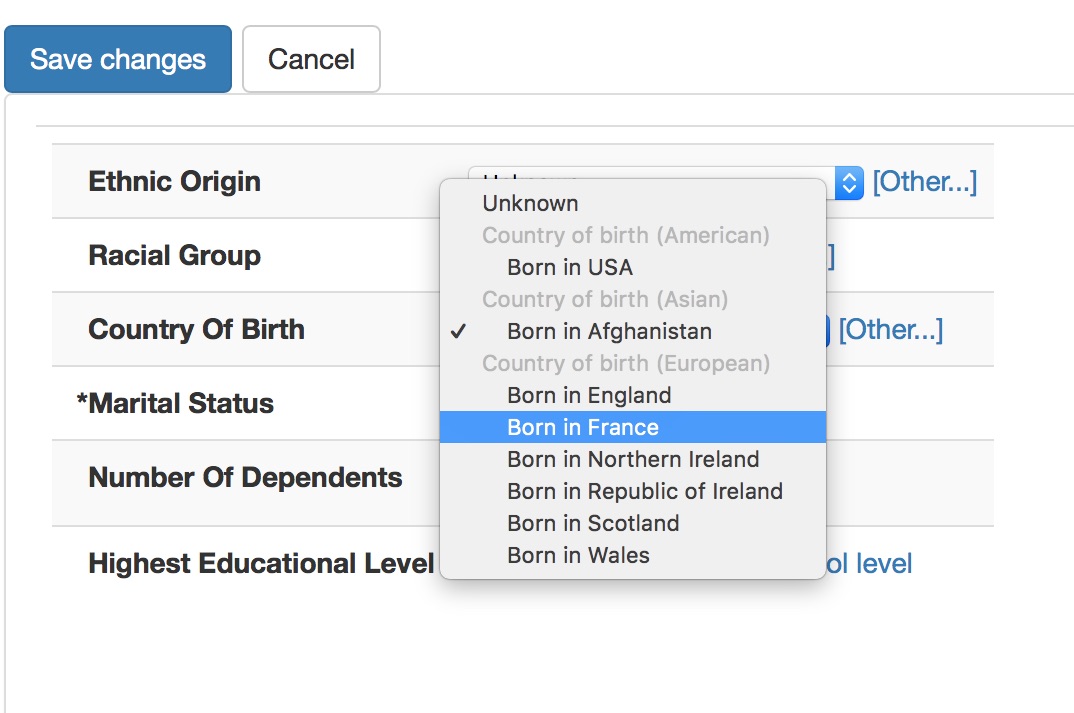
Notice how the countries are grouped as American, Asian and European. This isn’t hard-coded but instead I have set a single property to categorise each country-of-birth term by region in the SNOMED-CT hierarchy.
Internally, I have built a range of different UI components that are configured declaratively to allow choice of SNOMED concept from different subsets or parts of the hierarchy. Here are the rules defining the country of birth pop-up.
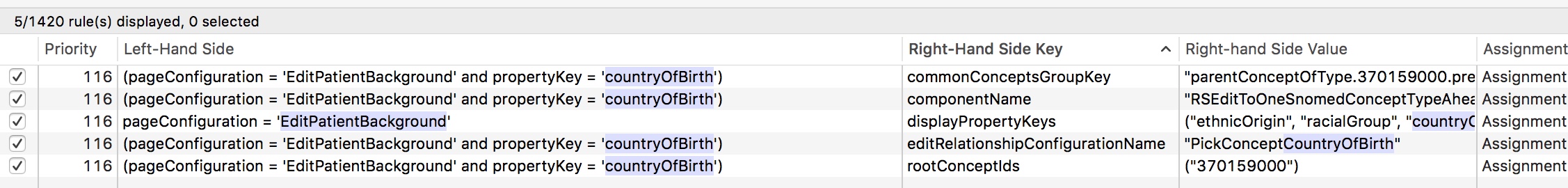
Declarative architectures are extremely powerful and support building systems responsive to need. I can change most of the application’s behaviour at runtime by using an administrative portal and the rule engine. Most data collection forms are automatically generated from the underlying data model without any additional work for me.
Entering terms should be quicker than using free text. Try entering “MI” into my basic demonstration application - https://msdata.org.uk/apps/WebObjects/SnomedBrowser.woa/
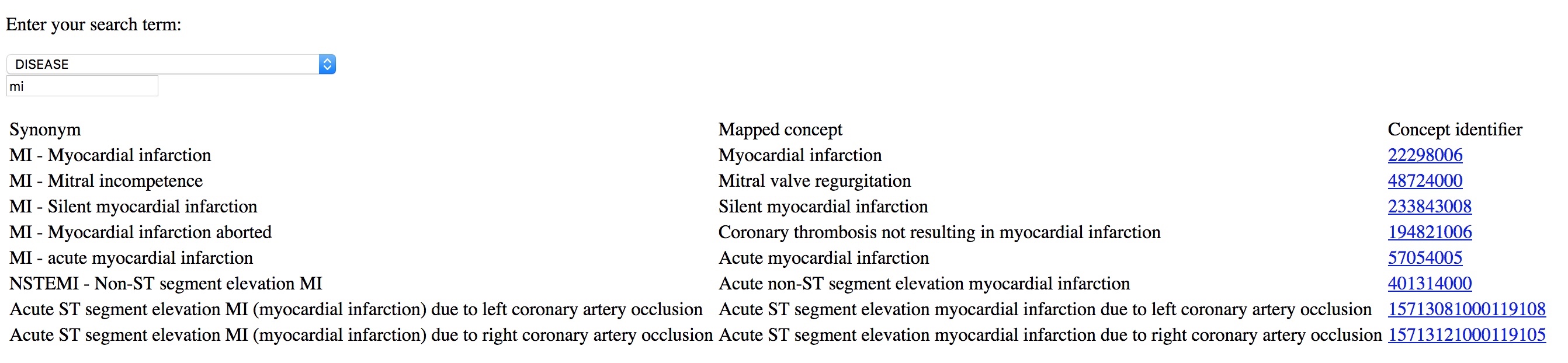
If you give tools to professionals that make it easier to enter coded structured information than free-text, they will do so. Coding diagnoses, procedures and background information is critical in understanding outcome data; they are a foundation on which more complex and sophisticated analytics can be built.
Patients
Since 2015, I have been trialling the use of iPads to allow patients to directly contribute to their records. The obvious next step is to roll this out more fully and permit patients to use their own devices, rather than devices we lend them in the clinic.
First, a member of the clinic staff logs in and chooses a patient from the clinic list for the day. This is fetched by the iPad via the EPR to one of two PAS in Wales to which I have interfaced, depending on clinic context. For demonstration purposes, I have built a fake PAS so I don’t inadvertently share patient-identifiable information for these videos:
The iPad is handed to the patient for them to complete. As the experience is locked to that patient, they cannot “go back” and look at the list of patients or see any information that they shouldn’t be able to see.
I can configure what to show dynamically based on diagnosis, context and previous responses. I want to start showing the complete patient record at some point.
So here is it running in “Parkinson’s clinic” mode, showing different questionnaires adaptively. The important feature is that patient data is available immediately for the consultation for clinical care.
In fact, most of the useful data goes into the automatically generated letter and is obviously available to be charted over time for that single patient, or aggregated later on for audit and research.
The patient is asked whether they wish to take part in research and then shown research projects and consent forms suitable to them, by virtue of applied inclusion and exclusion criteria, mainly using SNOMED-CT subsumption.
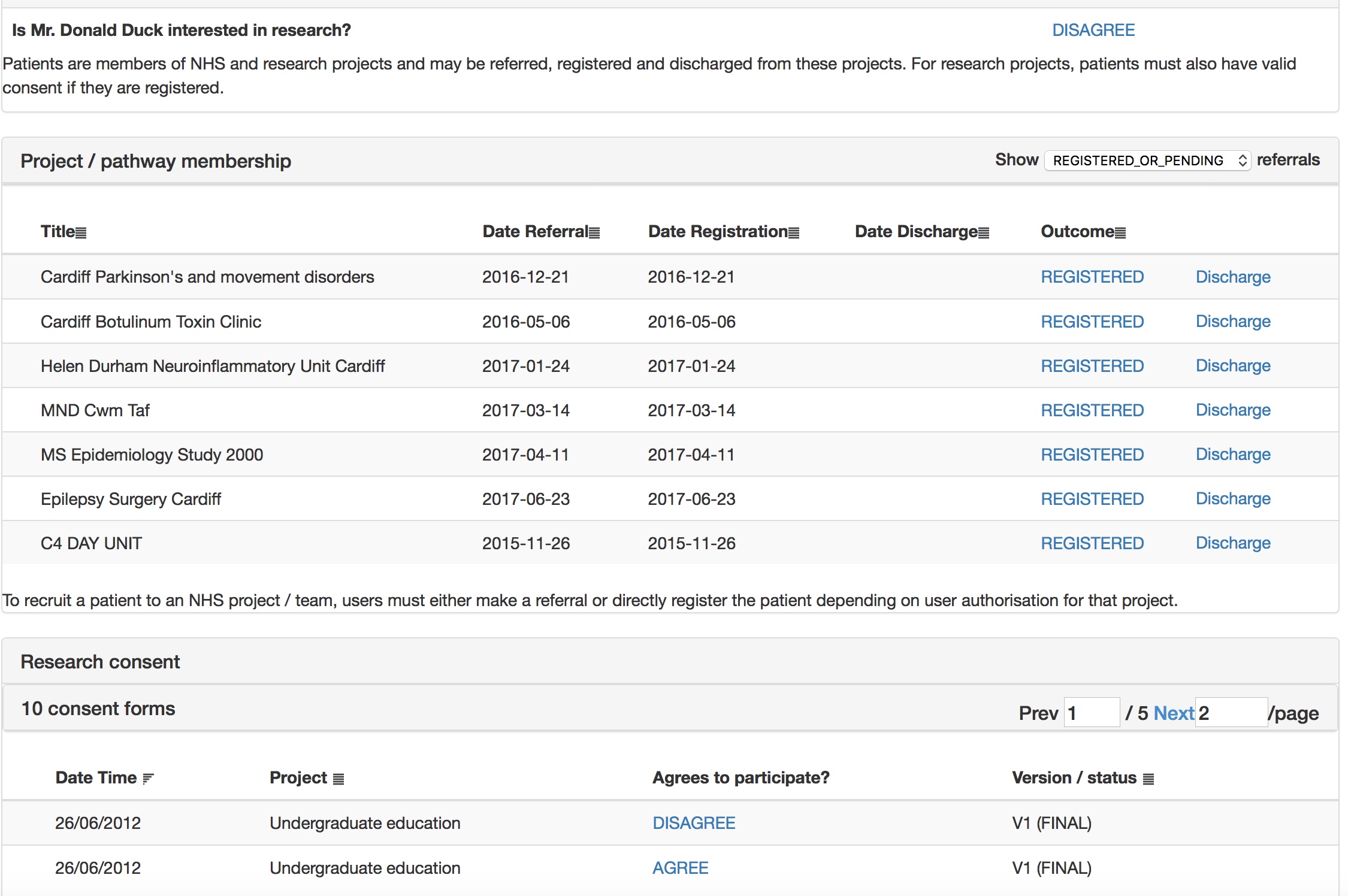
This is all configured at runtime obviously using my administrative portal. Here I edit the consent form interactively.
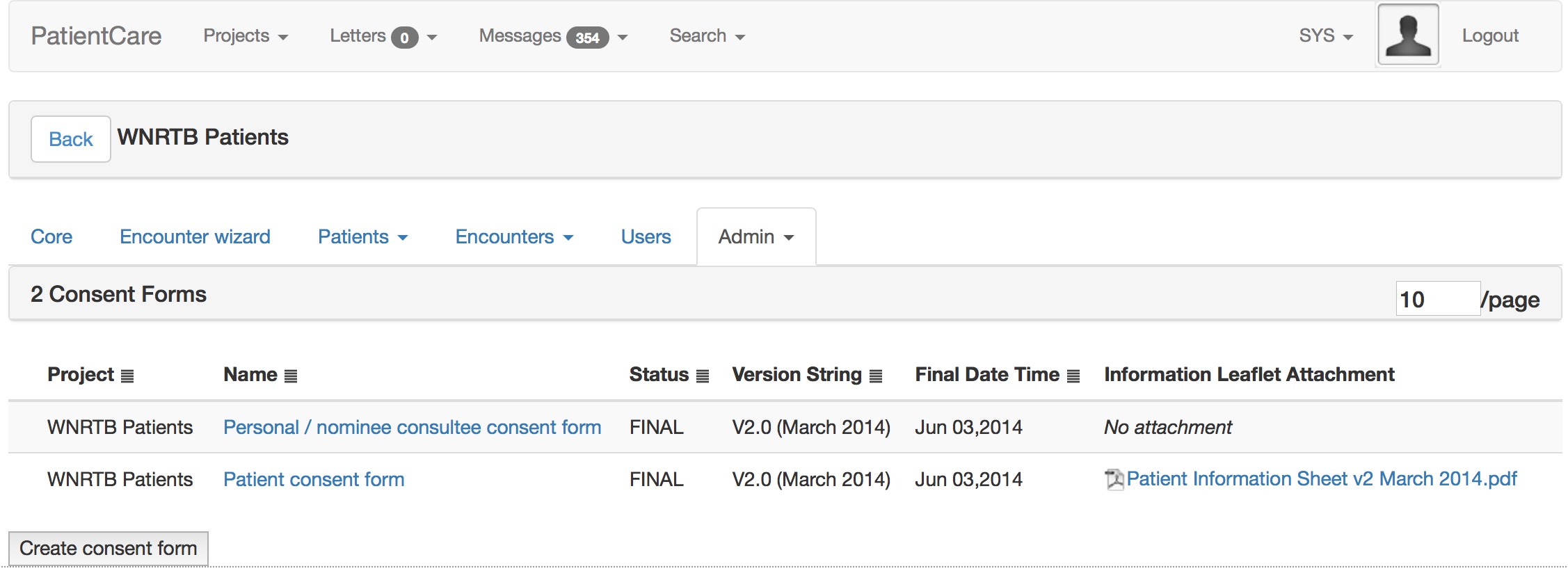
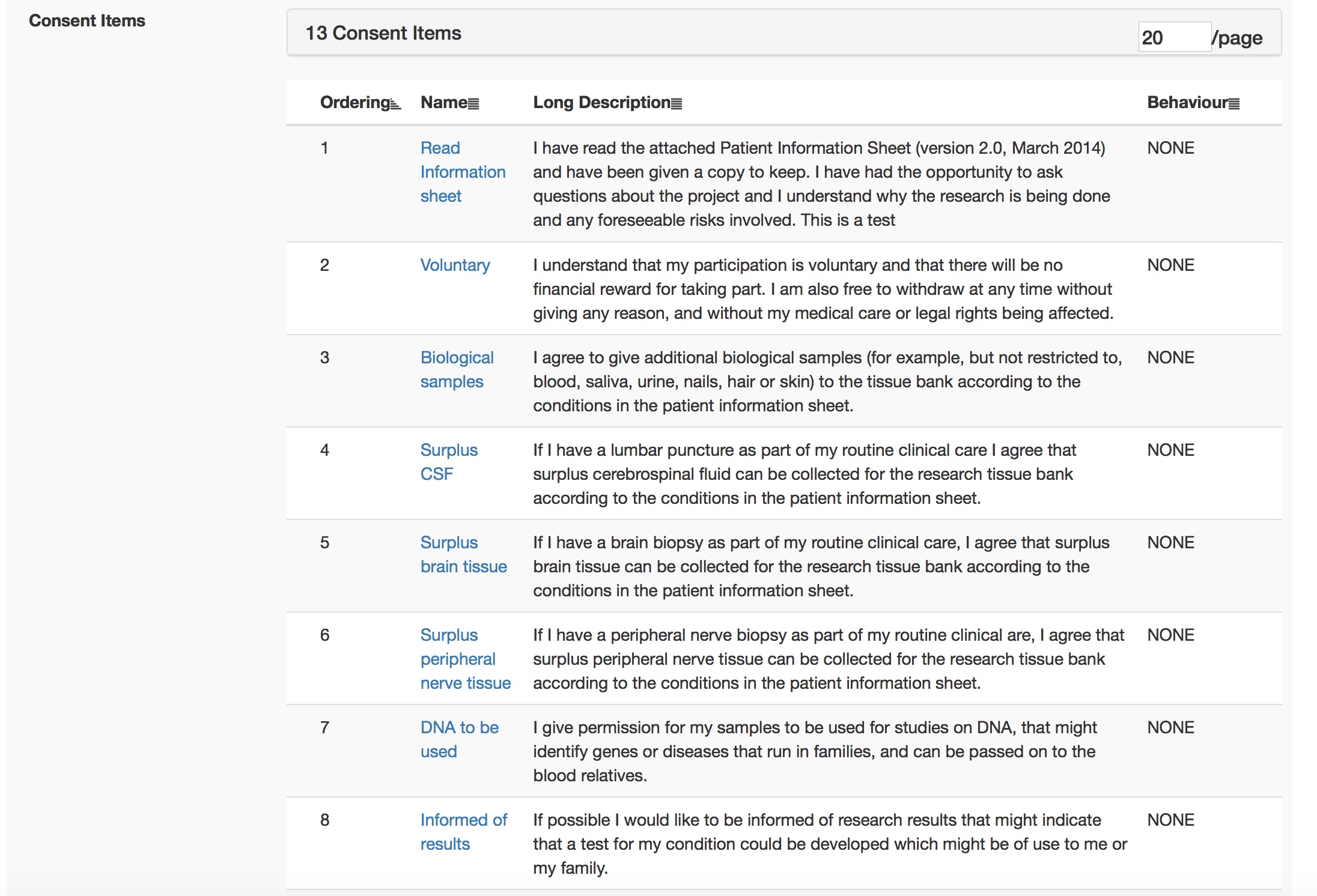
And once I sign off that consent form, it is used immediately by all iPads.
Technical architecture
I have built it slowly and incrementally using PostgreSQL and java for the web-based application. This exposes an interface for humans via a web front-end and a range of RESTful services for use by machines. I built the iOS applications in objective-C originally and then migrated to Swift for the current versions. These talk to the main EPR via HTTPS. The iOS applications use Apple’s ResearchKit framework to implement dynamic questionnaires and active tasks such as a finger tapping test.
One of the most important facets is the abstraction of multiple other external services, such as services provided by a hospital PAS (patient administrative system) and document repositories. While my software could be used standalone, it isn’t really designed to work like that and instead wants to interoperate with a broad set of external services. It wants to talk and exchange structured codified clinically-driven data with other systems.
So much software is focused on a single organisation, but our multidisciplinary teams cross both disciplinary and organisation divides! Therefore, my application can be configured to interact with multiple external software systems to make the user experience more seamless.
What has this taught me?
The benefits of:
- Technical and semantic interoperability with the use of standards
- Encapsulation and substitutability
- Decoupling
- Coherent architectural design
Much of the hard work has been in getting permission and then implementing technical interfaces to other systems. It would have been much easier for me to create valuable software if I had a set of tools and frameworks on which to build. This is reason why I am a advocate for an open platform approach in healthcare information technology so that we can build a stable infrastructure and yet still enable transformative innovation and embrace new digital technologies such as machine learning. I shouldn’t be wasting my time building the foundations of applications, but instead I should be able to leverage a suite of frameworks and tools on which hitherto unconsidered new and exciting functionality can be built.
When any of us build an application for iOS or Android, we don’t start with nothing but have a suite of frameworks and tools to enable drag-and-drop user interface design, data persistence and cloud synchronisation. Why don’t we have the same for healthcare yet?
My future plans are to break-up the application into a suite of interoperating microservices. I have already done that for my SNOMED-CT terminology engine, first of all as a java-based microservice and now one built using golang. As I do this, I rewrite my original application to make use of the standalone service.
I have written about the potential benefits of microservices and serverless architectures in healthcare previously), including the declarative nature of combining those services together by carefully considering how data flows between them in order to deliver value.
We need open architectures that interoperate, are standards-compliant and multi-layered. Foundational services, such as data persistence and core services like terminology should be used by more specialist and increasingly high-value functionality such as read-time analytics and aggregators.
I see a future in which those foundational services are provided via open-source tools and new commercial models evolve selling advanced services on a pay-as-you-go model. For example, would any of you want to take advantage of a disease-modifying treatment service that monitors your patients on drugs like methotrexate and orders blood tests when needed and lets you know when their blood counts drop?
We need to define a standards-based framework which means that we can not only deploy such innovative technology, but enable their assessment and evaluation in a safe way. One of the ways of enabling that are the principles of decoupling, encapsulation and substitutability, in which we can safely replace a modular component without other modules being aware. Only then can we enable safe technological-focused quality improvement and research, by running A-B testing in which different algorithms can be tested in isolation.
Such aspirations require a renewed focus on consent and in particular, making it possible to seek explicit opt-in consent in an easy way from citizens. Why not use technology to show informative video, interactive consent tools and start to recruit the majority of our patients into many different types of research project, obtaining robust and useful data on the effects of interventions, including the applied use of machine learning and algorithms, across our health systems?
We can no longer think in terms of monolithic applications and think of interoperability as something to tack on after those applications are finished. Instead, we should focus on how we build an open platform to enable distributed applications and services and permit innovators to stitch together that functionality in novel ways to support and improve and re-imagine our healthcare services.
Mark