Semantic interoperability: SNOMED CT, post-coordination and the model
Update 2021: You can find the latest version of my SNOMED CT library and microservice: hermes
One of our goals in healthcare information technology is to enable the meaningful exchange of information between distributed systems in the support of three broad purposes:
- direct care,
- management and quality improvement
- clinical research.
However, it is insufficient to simply exchange information at a technical level, but we must aim for semantic interoperability so that we exchange meaningful data.
When I say meaningful data, I mean data that has meaning and can be used for decisions, in order to reduce uncertainty about those decisions. Those decisions may be made by a professional, by a patient or carer, or much more likely, shared decisions among groups of those individuals. Those decisions may be clinical, such as should we start anticoagulation to prevent a stroke, or administrative, such as how many patients should we book into that clinic?
But when I use the term meaningful data, I also include the ability to use software to process that information; it is computable. It is important to understanding the benefits of computable information, because computer software is quite simply the best way to make sense of large amounts of information. Making data computable means our clinical work can become data-driven.
Making our data computable means that machines can help in processing and understanding, and with the advent of machine learning, even make or suggest decisions based on information. But this post isn’t about machine learning, although it is, I believe, a foundational step for the appropriate and meaningful development and application of machine learning in healthcare.
So this blog post is about how we can record information in a way that is useful to both humans and machines. For humans it should mean data can be entered more quickly and for machines, it permits better processing so that they can support us humans!
How to be data-driven
SNOMED CT is a comprehensive terminology and ontology for use in healthcare and it forms an important component of the solution to semantic interoperability. I implemented SNOMED CT in a electronic patient record system and it has been live and running for clinical use in the UK since 2009.
The SNOMED CT model consists of three core entities:
- concepts, represents a unique concept
- descriptions, represents synonyms for a concept
- relationships, represents relationships between concepts
I’ve written extensively about SNOMED-CT on this blog; the best way of understanding the ontology is to browse using an online browser but SNOMED International provide a useful introductory guide
Concepts
Many concepts represent a single independent concept, but many are actually compound, representing a number of concepts combined into a meaningful whole.
For example, cholecystectomy (38102005) is a single concept representing removal of the gallbladder. Laparoscopic cholecystectomy is single concept representing removal of the gallbladder with a laparoscope (45595009).
A SNOMED CT expression combines concepts without needing every possible combination of concepts to be created within the terminology. For example,
80146002|appendectomy|
:260870009|priority|=25876001|emergency|,
425391005|using access device|=86174004|laparoscope|
represents performing an appendicectomy as an emergency using an laparoscope. We could add this as a distinct single concept into SNOMED CT but that would result in a combinatorial explosion; that means we’d have to have single concepts representing all possible permutations which is simply not practical. However, mainly for historical reasons or to ease implementation, SNOMED CT does already contains many single concepts that combines multiple concepts into one.
However, SNOMED CT expressions are a powerful way of refining a concept with additional information, creating, in effect, a clinical phrase. This technique is called post-coordination.
Why not use post-coordination?
I have usually advocated keeping the implementation of SNOMED CT simple by using only pre-coordinated concepts. My concerns about using post-coordination have been that:
- it added complexity to end-users in terms of selection and display of terms,
- it made data storage more difficult, and would break referential integrity in my own system.
- it would complicate using those terms at a computational level e.g. in automated decision support.
- we gain significant value simply from recording pre-coordinated concepts
It is (1) and (3) that concerned me most.
For example, in my own electronic health record software, users don’t know that they are selecting SNOMED terms; they start typing and it autocompletes or simply choose an option using a pop-up, choose a part of the anatomy from a picture or a term is imputed on the basis of other data entered. Adding complexity in adding refinements or qualifiers or embedding expressions worried me. I wanted to make SNOMED CT invisible to end-users and yet enable new decision support and analytics capabilities, hitherto not possible using unstructured free-text.
I hadn’t conceived of a simple way to make using SNOMED invisible if I created functionality to build expressions; I now think differently.
Information models
However, in the majority of cases, I think many uses of post-coordination such as qualification or refinement or additional information should instead be a function of the wider information model in which the SNOMED CT concept is recorded.
For example, the FHIR definition of a Condition, which includes metadata about the problem/diagnosis concept is shown here:
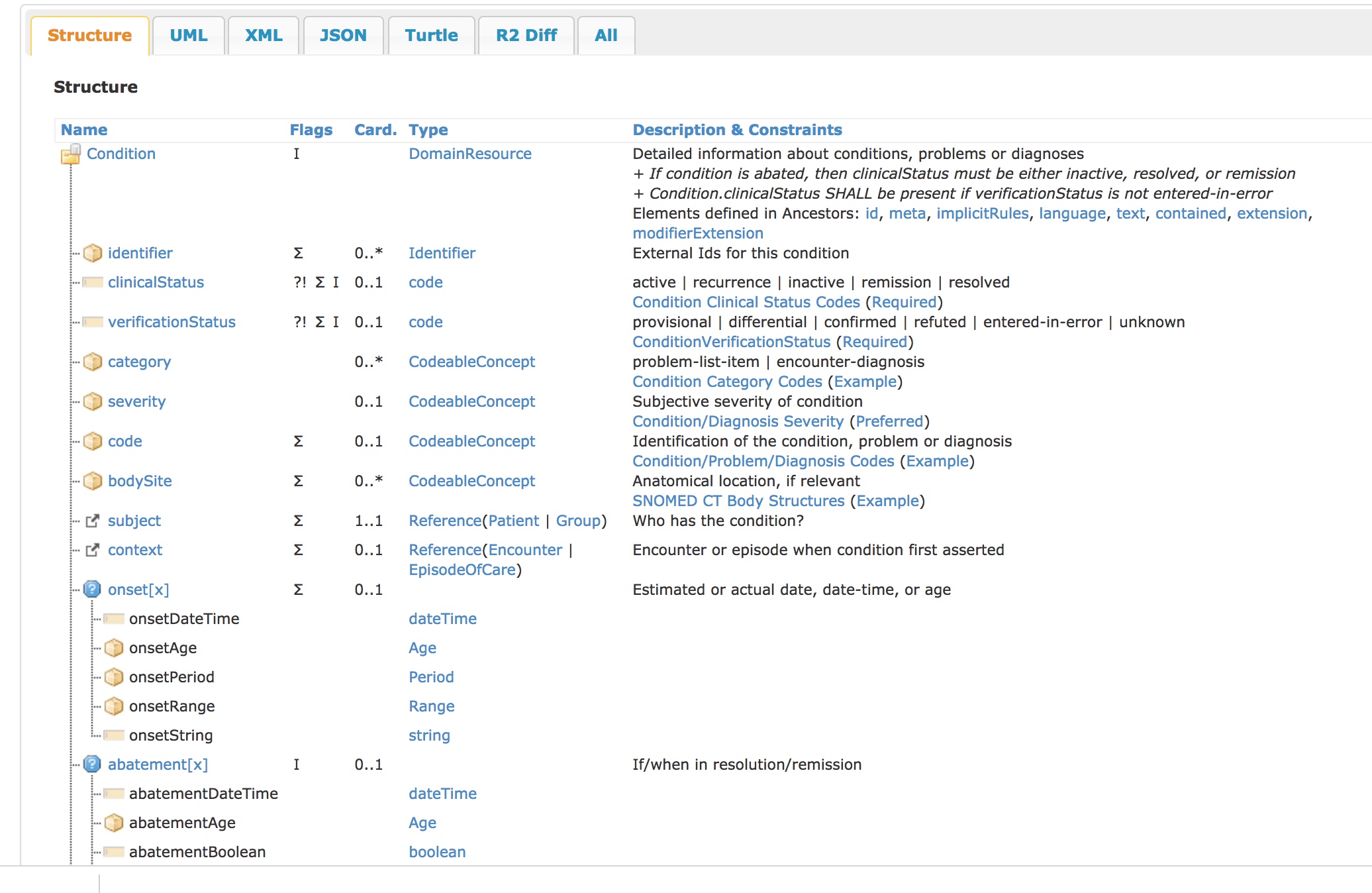
It includes information relating to status (active/inactive etc), verification (provisional, differential) and severity (severe, moderate, mild), as well as information about the condition’s onset and abatement (remission/resolution).
If we use post-coordination without carefully considering the consequences and their mitigation, given that there is an overlap between information recorded within an expression and the wider information model, we risk duplication, complexity and inconsistency.
Computing attributes
But there are clearly situations where refining a concept with additional information would be helpful. For example, we really wanted to be able to add a more specific anatomical location when we recorded that a patient had a brain abscess.
The SNOMED-CT ontology helps us, at the computational level, determine how “cerebral abscess” can be refined:
A cerebral abscess (60404007) is an abscess in the brain.
Firstly, we can determine a way to refine this concept using post-coordination to specifically give the location. The ontology provides, as part of its model, attribute relationships for this concept, at a computational level:
Is a (attribute) -> Abscess of brain (disorder)
Associated morphology (attribute) -> Abscess (morphologic abnormality)
Finding site (attribute) -> Cerebral structure (body structure)
For example, we can see that we can refine the concept cerebral abscess with a finding site that is a child of the concept Cerebral structure (body structure) (83678007).
As a result, computationally, we can offer a choice to the end-user to refine this concept with any finding site, such as Structure of parahippocampal gyrus (13989007)
Secondly, we can add refinements as a result of our concept being in one of the nine main SNOMED CT hierarchies. The attributes appropriate for each hierarchy are listed on the SNOMED International concept model documentation. For example, a clinical finding can be qualified by adding concepts representing a Finding Site, a Causative Agent, or Severity among others.
Thirdly, we can add refinements because of the existence of our concept within a certain reference set. For example, there is a laterality reference set that tells us which concepts can be lateralised.
It is therefore straightforward to imagine that, if a cerebral abscess is entered by a user as a diagnostic concept, we might configure our user interface or natural language parser to look for and refine that diagnostic concept with additional information relating to finding site, appropriately limited to the brain using subsumption, given that we have entered cerebral abscess, and offering a choice of laterality.
So we can see that location, laterality and causative agents might be very appropriate options for refinement, but should we let users post-coordinate diagnostic concepts with a type of ```Severity’’’? This matters because we need to think about how we write software to process these terms.
Equivalence and the importance of meaning
The key issue with processing SNOMED CT, either as pre-coordinated terms or as expressions, is to ensure that we can process meaning.
That means that, in any implementation of SNOMED CT, whether using only pre-coordinated terms or post-coordination to create an expression, we need to handle equivalence at not just the level of SNOMED, but also take into account the wider information model, providing the context in which that concept or expression sits.
For example, a HL7 FHIR representation of a condition looks like this:
{
"resourceType" : "Condition",
// from Resource: id, meta, implicitRules, and language
// from DomainResource: text, contained, extension, and modifierExtension
"identifier" : [{ Identifier }], // External Ids for this condition
"clinicalStatus" : "<code>", // C? active | recurrence | inactive | remission | resolved
"verificationStatus" : "<code>", // C? provisional | differential | confirmed | refuted | entered-in-error | unknown
"category" : [{ CodeableConcept }], // problem-list-item | encounter-diagnosis
"severity" : { CodeableConcept }, // Subjective severity of condition
"code" : { CodeableConcept }, // Identification of the condition, problem or diagnosis
"bodySite" : [{ CodeableConcept }], // Anatomical location, if relevant
"subject" : { Reference(Patient|Group) }, // R! Who has the condition?
"context" : { Reference(Encounter|EpisodeOfCare) }, // Encounter or episode when condition first ...
}
A CodeableConcept is a complex HL7 FHIR type that provides a reference to an arbitrary terminology.
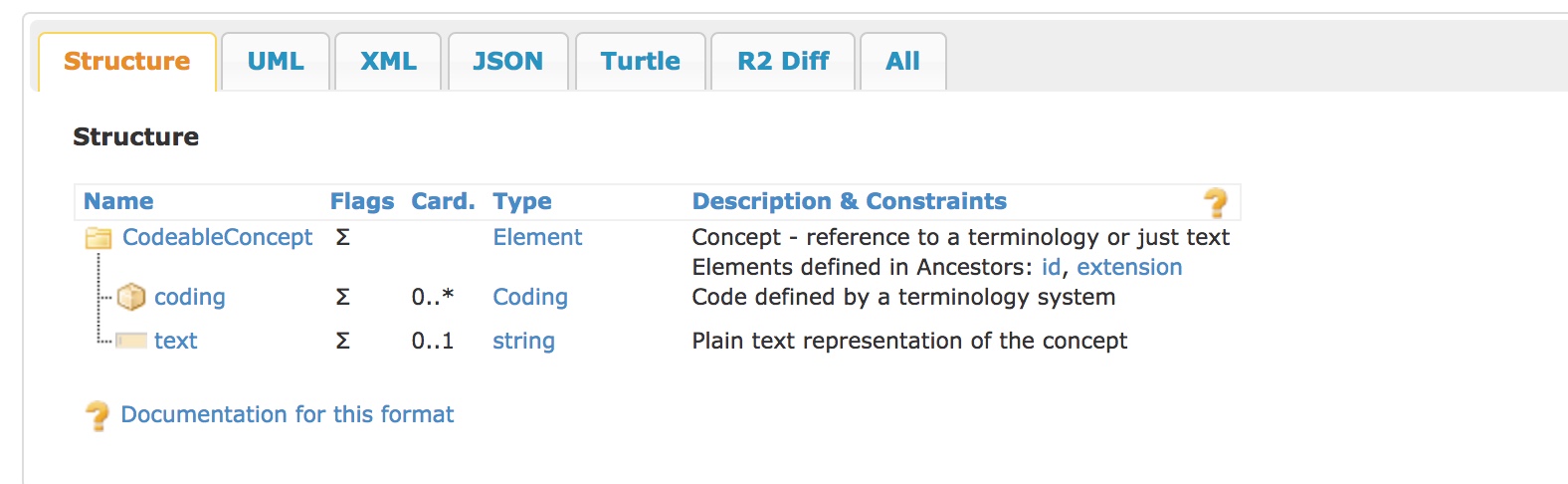
What happens if we put a concept representing “SEVERE” as the severity in the FHIR model but record a post-coordinated SNOMED CT expression of “MILD” as a qualifier of our diagnostic concept?
Meaningful data, in context
That means our analytics, for direct individual care, processing data as part of an electronic health record in real-time, or for quality improvement, service management or research use, must interpret the meaning of SNOMED CT concepts and expressions within the wider information model.
So what do we need from our software?
-
We need to be able to process an arbitrary SNOMED concepts, provide the standard attribute relationships to client software so that those developers can easily understand how to refine, for example, a diagnostic term. This should include the value sets or necessary search constraints so that only appropriate values can be used for any specific attribute (e.g. if you are refining an anatomical location in the central nervous system, your values should only be anatomical locations within the central nervous system!)
-
We need a set of tools that can parse SNOMED expressions into a useful internal representation and vice versa, so that expressions can be persisted and exchanged as well as permitting the information to be used effectively by humans and machines.
-
We need to be able to process an arbitrary SNOMED concept or expression and normalise it into a canonical expression so that it can be used for analytics.
-
We need tools to be able to offer additional refinement of SNOMED concepts and expressions by virtue of the wider information model in which they sit. For example, normalisation of a concept within a FHIR condition model could be automatically refined by the appropriate attributes from the wider model, such as severity.
As a result, I have been working on a set of open-source tools that not only support deriving understanding of meaning from pre-coordinated single terms in isolation, but from SNOMED expressions and those terms in their wider context.
As part of this work, I have made use of the Google HL7 FHIR protobuf definitions, which can be found here and are now vendored in the go-terminology code repository.
My next blog post in this series will cover the details of a practical implementation. Like all standards, it is only with the development and availability of quality (open-source) implementations that a standard can truly become valuable.
Mark