The semantic patient
I’ve been off work, off blogging and off Twitter due to COVID-19 but fortunately now back to full health and thought I’d write up some notes about what I’ve been doing in my spare time outside of my day job doing telephone clinics and COVID ward duties.
Lightweight applications
The first is experimenting with lightweight mobile and web-based applications that provide a compelling way for users to interact with health and care data. Separating applications from the data is important, and web-based technologies have matured sufficiently so that one can build a desktop-class user experience in the browser.
I built a proof-of-concept system to record and display electronic observations (pulse, blood pressure etc) and calculate national early warning scores for escalation. It integrates with existing NHS systems for user login and patient lookup and was about three days work I guess, but spread out over a few weeks. It dynamically generates an SVG NEWS chart, which looks pretty on iPads or on whiteboards and it was my first ever clojure application. It’s all dynamic and one can scroll to view results over time, and I’ve built SNOMED CT autocompletion widgets and am planning ward round problem lists and actions/ordering.
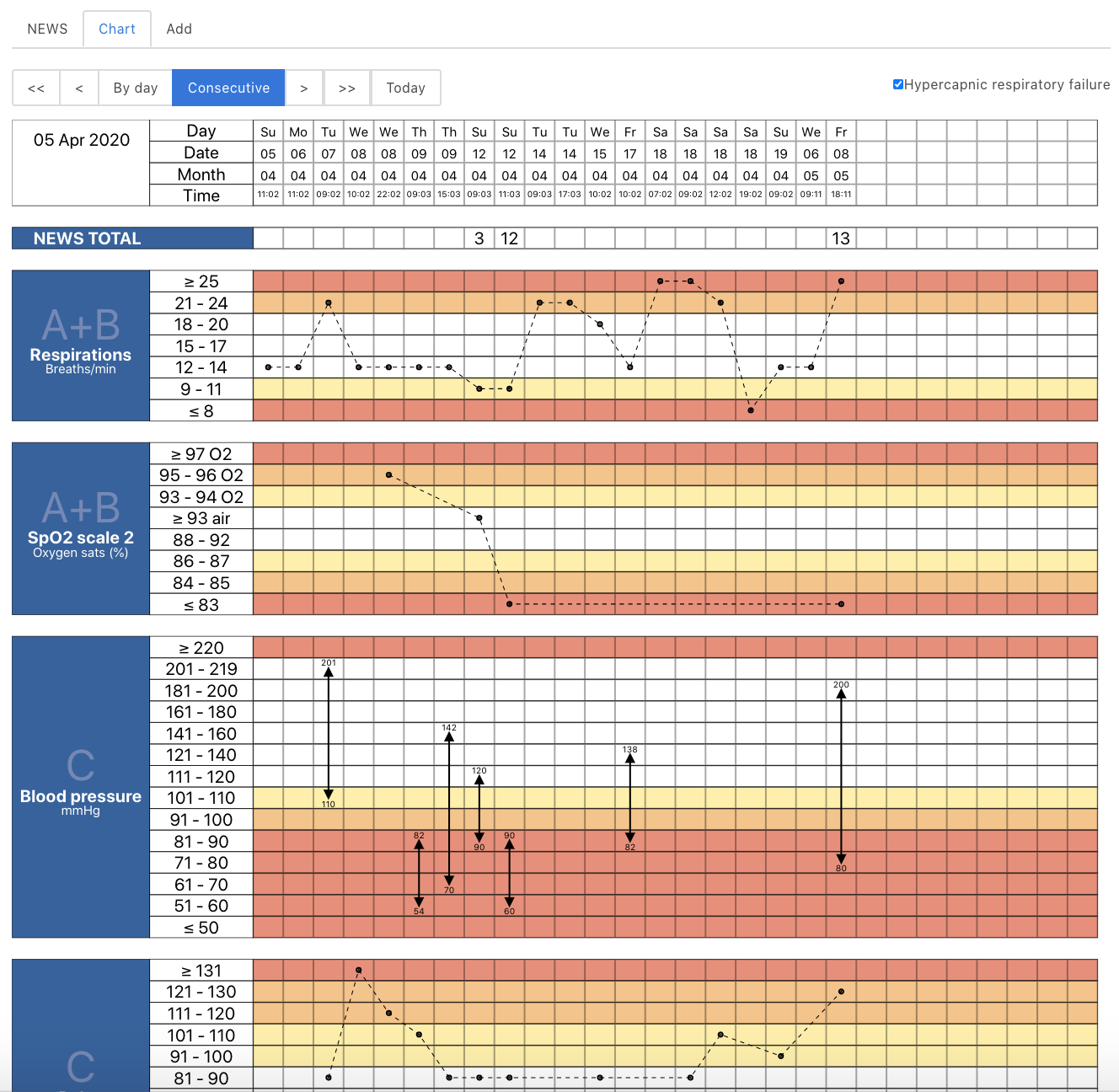
You can also see a video of the application running.
The source code is open as well.
Data, dynamism and ontologies in health and care
But my main health informatics work has been about linked and self-describing data.
Our ability to write software is critical in building a learning health system, but it is very easy to build rigid, hierarchical, poorly adaptable and brittle systems - and that is normally the result if one tries to solve problems through the lens of a single application or portal, instead of focusing on data and an architecture that enables the recording and processing of those data in order to inform care.
We must strive to build health and care systems without walls, and create effective tools for health and care organisations, and citizens, to manage, interact, plan and undertake care. Technology is critical in creating health services that work safely and effectively for patients and their carers. In banking we see technology companies entering the industry, and established banks trying to become technology companies; they have recognised the critical need to harness technology to improve their services.
“Health and care needs to learn how to deliver software : reliably, safely, reproducibly, at scale and to ensure ongoing monitoring, evaluation and continuous improvement.”
https://wardle.org/strategy/2019/09/22/health-technology.html
We should be building a technical architecture for health and care that is safe, responsive and effective. We must look at how we can stream data independent of individual applications and make use of real-time analytics and advances in technology to best inform and improve care, both for an individual and cohorts of citizens. We cannot achieve our goals unless we think about health and care as it truly is, a symphonic orchestra with many players, all needing the right information at the right time, and all working together in concert. They play different instruments, and have different roles, but they work together.
It is naive to think that we can have the same software running in critical care, on a patient’s own devices, in a nursing home, in the outpatient clinic, whether virtual or in-person, in the emergency department, in the GP surgery, in surgical theatres or on the ward. User-facing applications must be focused on improving process and workflow and so are absolutely dependent on the clinical context in which they will operate. There are commonalities, of course, but truing to make change using top-down, command and control management and multi-million pound programmes is not the way to improve the working of complex adaptive systems. We have only started the journey on how to make computers effective in medicine.
One of the best ways of making valuable software is to break-down the problems we are trying to solve into smaller problems, each of which one person, or one team can conceive of and solve in one go. It’s one of the reasons why I like domain-driven design and understanding the importance of scope, so that I can focus on the problem at hand, and decouple the component parts of the wider system.
A logical consequence of such an approach is of abstraction so that the other parts with which we interact are abstract; I don’t need to know how you are storing that clinical document or exactly what happens after I send those data but I do need to have a set of contracts, covering the technical, legal and regulatory matters that define our interactions. In essence, those contracts, including the technical standards that define how those interactions proceed, can be a powerful force for speeding up our developments which can now happen in parallel and limit the need for constant interaction between teams.
But abstraction also should extend to the meaningful data that we so depend upon to care for individual patients, manage and improve our services, and continuously improve and learn.
Having meaningful data means that we can make sense of that data - we can derive its meaning. It therefore follows that not only do we need to be able to exchange information at a technical level, via technical interoperability, but we need shared understanding of meaning via semantic interoperability.
That’s not to say exchanging unstructured documents isn’t valuable. It is, but it only goes so far. I can’t do much as much as I’d like with a binary PDF document.
The problem with health and care data is that is hard to interpret out of context. If your pulse is 150/minute, is that a problem? Yes if you are asleep but no if you are running or playing tennis!
And that’s why health and care data needs context in order to make data meaningful. We need a shared understanding of a wider information model.
Those measurements such as pulse, blood pressure and pain scores need to anchored within a context - my neurology clinic for that new patient appointment in that clinic suite, in that unit, in that site, for that organisation, as part of that clinical pathway.
Meaningful data, using persistent meaningful identifiers
But how do we represent health and care data when it is complex and ever-changing? New diagnostic tests and new diagnostic entities are added to the corpus of our knowledge all of the time, and previously known “facts” have changed or replaced with our ever greater understanding?
But such changes don’t only apply to medical knowledge, but also apply to the very organisations within which we work! I used to work for Cardiff and Vale NHS Trust, but that organisation no longer exists and instead I work for Cardiff and Vale University Local Health Board! A core trainee will be a consultant in a few years time. Change is constant.
Here is a screenshot from one of the NHS Wales’ national portal (there are multiple instances each which gives the same view of sometimes different data). It provides a faceted search across documents, by specialty, but because the team didn’t adopt any standard for the coding of specialty, “Neurology” is different to “NEUROLOGY” and “NEUROLOGY EPILEPSY” its own specialty but isn’t included in either of the other two.
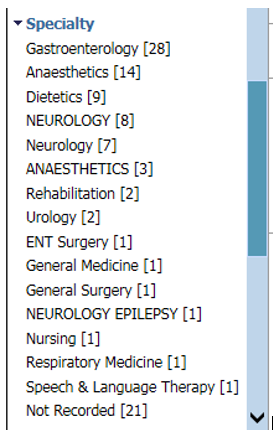
When I’m trying to care for a patient, I have to laboriously click on each in turn in order to find the information I want, simply because the designers of the service did not pick a useful vocabulary of terms to which documents could be tagged.
One of the ways we suggested avoiding this situation right at the beginning was to use an ontology to describe the specialty, such as SNOMED-CT, which provides a flexible, hierarchical and nested structure so that, for example, the epilepsy subspecialty is actually a subtype of the wider neurological subspecialty. That would have meant we could let users filter for “Neurology” and actually include subspecialties such as “paediatric neurology” without any great computational difficulty. I’m hoping this application will adopt this, although it seems to have taken many years and still it hasn’t changed as of today at least. I live in hope.
SNOMED CT, like many other ontologies, has multiple hierarchies, which means that it can model complex nesting, such as ‘bacterial meningitis’ being both a ‘neurological disease’ and an ‘infectious disease’. In addition, its identifiers are persistent. Even if ‘Wegener’s Granulomatosis’ is no longer the preferred label for that disease entity, the model explicitly manages change over time. There are mechanisms for making change, although it is a great shame that the distribution is not published under an open licence.
This idea of persistent identifiers is not new but such an approach is essential when we are dealing with decentralised, decoupled systems in which we need to record a reference to information in another system. You can think of persistent identifiers as analogous to ‘foreign keys’ in a relational database table.
In ontologies, we can also relate one identifier with another as a ‘triple’. For example, in SNOMED CT, we have
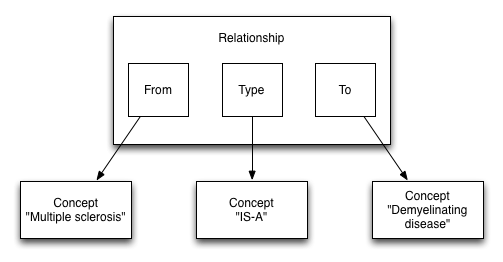
We can see that these triples help us build a directed graph that we can navigate and answer questions like, “Is this disorder a neurological disorder?” without difficulty.
Commonly, we can use identifiers that are a tuple of a namespace and an identifier. For example, we can create a SNOMED CT expression using “http://snomed.info/sct” and “24700007”, which together represent the concept of ‘multiple sclerosis’ and can therefore be computed as such.
HL7 have been doing this for years, with “object identifiers” or OIDS. In fact, the HL7 UK OID for representing health and care organisations uses the OID “2.16.840.1.113883.2.1.3.2.4.18.48” and these OIDs are published via Organisational Data Services (ODS) API endpoint.
Here is a data representation of the University Hospital of Wales: https://directory.spineservices.nhs.uk/ORD/2-0-0/organisations/7A4BV
HL7 FHIR uses this identifier model extensively; its model contains a namespace and a value, together with some additional metadata, which in hindsight should be a function of the resolution of that identifier rather than in what is essentially a foreign key.
Namespaces can be arbitrary values such as an OID, or a URI (uniform resource identifier), and while useful, much like an ISBN for a book, they do not actually provide a way to identify the location of the resource that they refer to. Resolution of the value within that namespace is left to you.
Namespaces can also be actual URLs and so can be resolved simply by accessing whatever information can be found at that web address, but this isn’t always the case!
Likewise, the same resource, such an organisation, might be represented by a number of different, and yet by definition, equivalent, identifiers.
For example, the University Hospital of Wales might be represented by
-
https://directory.spineservices.nhs.uk/ORD/2-0-0/organisations 7A4BV -
2.16.840.1.113883.2.1.3.2.4.18.48 7A4BV -
urn:oid:2.16.840.1.113883.2.1.3.2.4.18.48 7A4BV -
urn:oid:2.16.840.1.113883.2.1.3.2.4.18.48 RWMBV
Similarly, some identifiers in truly different namespaces actually point to the same ‘concept’, whether that be a diagnostic term (e.g. Read code for multiple sclerosis is, semantically, the same as a SNOMED CT code for multiple sclerosis) or a staff identifier should resolve, at some level, to the same ‘person’ as their GMC code.
Resolution, mapping and the testing of equivalence of identifiers is therefore a first-class problem in our domain, and should be modelled, and managed as such.
Dynamic and federated resolution of identifiers
We can overcome the challenge of identifier resolution, mapping and equivalence by making identifier namespaces an ontology by providing open data and open tools in order to ‘make sense’ of arbitrary identifiers within our domain and providing dynamic, flexible and adaptable mechanisms to extend and federate. This is akin to the domain name service ‘DNS’ of the Internet, which maps arbitrary textual domain names to their IP addresses.
These ideas led to me to build concierge, a set of simple ‘identifier resolvers’ that cover a set of practical examples - for my case, resolution of staff and patient identity, organisational data services and terminologies. The source code is available on github: https://github.com/wardle/concierge. I used it in my e-observation toy demonstration above for user login, fetching staff information (name, job title etc) and patient search/lookup. It is incomplete, but a useful exercise. I can now conceive of a single ‘Patient’ resource that uses information from multiple sources such as local patient administrative systems and the national enterprise master patient index to generate a unified contact telephone list, for example. I plan to abstract federation, so that client applications can start from an organisation, drill down to a site, and then drill down even further onto units, wards and even individual beds, with data derived from different systems for the different levels of granularity, but that complexity is hidden from the client user, and client application acting on their behalf.
Similarly, it is easy to consider how a single “staff” search/identifier resolution service might work to federate and abstract information from multiple disparate sources ; the staff directory, the electronic staff record, the on-call rotas, team definitions etc. There are many different ways to federate; the focus is to abstract underlying complexity for clients of that service.
But what about shared models?
So it’s easy to see how we might create a tidy facade to hide the underlying complexity of our existing health and care infrastructure using first-class identifiers for different resources. For example, we could resolve and reify a staff member and that data model could identifiers representing the organisations and the sites in which they work, or the clinics for which they are responsible, which themselves are linked to organisations and sites. Indeed, it’s easy to see those relationships as forming a graph, not only within a single ontology but between ontologies, which I’ll discuss shortly.
The problem is that not only do we need a shared understanding of identifiers, but we also need a shared understanding of the data models in which they are used. It is not sufficient to have a flat list of other identifiers within, say, a member of staff’s record because that would be meaningless; we need more context about how one relates to another.
Answering the question “does this patient have a type of neurological disease?” and getting back an answer “yes” also means that we need to be able to resolve an identifier in context in order to derive meaning, because that diagnosis of “Parkinson’s disease” might be listed as part of a family history and not in the patient’s personal problem list.
Others have solved this in different ways, and have even used different approaches within the same wider model. For example, the HL7 FHIR “Patient” model defines a type of resource representing a patient, with attributes such as name, gender and birth date. HL7 uses a more flexible, dynamic system for other models based on identifiers such as that for observations. Here is an example for recording glucose which, rather than hard-coding the test and the value of the result, uses namespaced codes and flexible values with coding for units. We can see that in general, the class is given prominence (Patient class, Observation class) and the attributes serve that class.
Likewise, in openEHR, named archetypes such as pulse and blood pressure can be composed into wider models for operational use, and these archetypes fit into a wider and more fixed information model as part of two-level modelling. For example, the blood pressure archetype contains a value for systolic blood pressure and a value for diastolic blood pressure, together with some metadata.
Neither the HL7 FHIR or openEHR approaches avoid the need for close collaboration between disparate health economies because unless one is not planning on actually using the data stored within the models, at some point one needs to write code that can consume and process and extract meaning from those data. In general, software developers compile those data definitions into classes to permit that processing.
If you’re going to exchange medication data, at some point you need to come to a decision about a shared model for recording, storing and exchanging that information, whether that exchange is simply from a server to a client or between a critical care software system and the hospital pharmacy.
Class-orientated vs attribute-orientated
In object-orientated software systems, data are usually coerced into objects of certain classes, and in most computer languages, those classes can be hierarchical so that for example a ‘Car’ ‘is-a’ type of ‘Vehicle’. There is usually a pattern of inheritance so that behaviour and sometimes data can be inherited from the parent class. For example, all vehicles might have a ‘colour’ property.
It’s a seductive approach, as one can build a model of reality and use it to solve problems by modelling data and behaviour as interactions between objects. You think in terms of objects, those objects may have properties, but you almost always have to define how those objects fit into your wider hierarchy when you create them. The hierarchies of your object classes are usually quite fixed.
Both openEHR and HL7 FHIR use a class-orientated approach to most of their data.
It’s seductive, but it can be brittle. It is generally poorly extensible, poorly dynamic and struggles to adapt not only to changing circumstances but even to different applications across a wider health enterprise, In some cases, these aggregates of attributes are grouped only for context and not for semantics.
HL7 FHIR adopts a modelling approach in which the basic class definitions aim for a common denominator of fields and permit “extensions” for local definitions while using profiling to make very general models more specific by, for example, defining what can be in a value set or which terminology should be used for a specific property.
openEHR adopts a different approach in which modellers attempt to define all of the possible attributes within the model, and use optionality extensively; for example it would be unusual to use all of the attributes in many models.
HL7 FHIR is therefore a ‘meta-standard’ and what really happens is that we as a community build multiple versions of the same ‘standard’ - see the US CORE release and there is proliferation of just-slightly-different profiles for different scenarios within NHS England.
Promoting first-class, namespaced attributes
But haven’t we been talking about ontologies in which we can associate arbitrary properties in triples, and change those over time? For example, work on the ‘semantic’ web led to the development of RDF (Resource Description Framework):
“RDF is a standard model for data interchange on the Web. RDF has features that facilitate data merging even if the underlying schemas differ, and it specifically supports the evolution of schemas over time without requiring all the data consumers to be changed.”
Now people have been talking about using RDF in healthcare for many years, but such an approach has not gained traction for a variety of reasons, including inadequate open-source tooling, little industry interest, perceived complexity, ownership of different ontologies by different groups, its design predating interest in graph databases, lack of definitions on value types and probably most importantly, a lack of temporal associations (timestamping) and change semantics.
But if we can solve those issues, it is quite easy to see how we can compose first-class attributes together into a semantic whole to represent health data in context. Just as in modern, more functional programming languages, elevating properties to first-class concepts within our models gives us powerful runtime polymorphism and permits independent extensions of types, protocols and implementations within and across health data.
But such an approach requires several important dependencies:
- Robust shared tools for the resolution, mapping and logical comparison/equivalence testing of persistent identifiers
- A move away from CRUD (create-read-update-delete) semantics towards different semantics for write and read operations. This has important regulatory benefits as well.
- A move to architectural designs that focus on immutable data and change as a first-class part of the model
- Denormalisation with snapshots of the dereferenced values and time as a first-class attribute, so that our software doesn’t show data recorded by me in 1999 using my current job title but my job title at that time.
- Clinical modelling using finely-grained archetypes representing a single attribute, properly namespaced and dynamically grouped into other less granular archetypes.
- Tools to provide automatic coercion of legacy “object-orientated” classes into bundles of first-class attributes.
- Ontologies describing the relationships between properties, even between different ontologies.
Such an approach also means we can support interoperability between different information models in our software systems, and build automated tools that take data from different systems and generate context-appropriate semantic data. In this world, we could make
https://openehr.org/Id/observation/blood_pressure/v2 systolic
equivalent to
http://snomed.info/sct 271649006
and equivalent to
http://loinc.org 8480-6
Unfortunately, the fact that the latter two are proprietary and need licensing is to our collective detriment, and many operational openEHR implementations are closed-source.
But it is easy to see how a class-based model results in an explosion of classes. For example, it’s not difficult to see how the first line of an address in a FHIR address class is semantically the same in a “UK” address from openEHR which is the same as in a Scottish address from openEHR which is the same as in an ISO22220 based address (also from openEHR - https://ckm.openehr.org/ckm/archetypes/1013.1.484 and all share the semantics of the vCard ontology ‘address’ even if the attributes might have different names.
Instead, of this duplication of classes, we can define ontologies that define the relationships between those first-class names and provide tools to arbitrarily resolve those identifiers. That means resolving “http://www.w3.org/2006/vcard/ns#street-address” can return the right semantic data irrespective of whether the source information is in HL7 v2, HL7 FHIR, or one of the many openEHR address archetypes. Ontologies can be used to federate attributes from different models.
One can write software to process the fully open openEHR archetype definitions and bind properties to first-class namespaced attributes and many definitions already have bindings to other definitions in LOINC or SNOMED CT. Both the openEHR and HL7 FHIR communities have done some great modelling work, including documenting the semantics of their types and attributes, and its possible to build tools to process those definitions.
Grouping first-class attributes into semantically meaningful entities
Just because we make attributes first-class in our models, it doesn’t mean that we can’t bundle them together into something that is itself meaningful. But rather than thinking in terms of classes, and the attributes are simply part of those classes, our attributes are first-class and might be arbitrarily grouped into what are essentially nested collections and be derived either directly from source information or derived by understanding how those properties relate to one another semantically. Grouping is based on semantics and not necessarily only contextual differences.
In addition, our definitions of an attribute must be associated with a schema definition, so that we have information about the value type(s) permitted for that property. Other semantics might include cardinality and uniqueness, for example, depending on the property in question. But object-orientated approaches conflate the specifications of data with the data itself, while in reality these are orthogonal concepts. Those specifications define the overall “shape” of our data.
It’s fairly easy to see how schema definitions could themselves be defined ontologically, through entity/attribute/value (or subject/predicate/object triples in RDF parlance) and so permit runtime evolution of health data schemas and importantly, an ability to provide a unified view from data sourced from multiple disparate systems. It provides a true abstraction and could present data from HL7 FHIR, HL7v2 and openEHR in the same way and so permit computability across different systems, while also providing a persistence mechanism via an entity-attribute-value store for native applications and services, and a way of generating computable properties in a functional style.
Hierarchical databases, graphs and graph queries
It is unsurprising that early medical databases, some still used today, model healthcare data in a hierarchy. What this means is that a single patient has multiple encounters, each encounter has multiple procedures etc. In relational database terms, each is a to-many relation.
However a simple hierarchy is insufficient for most health and care needs. For example, a clinical encounter might relate to multiple pathways of care, in a many-to-many relationship. Instead, we can picture the modern medical record as a graph of information with our ‘triples’ forming the nodes of that graph. Some nodes will be highly connected, others less so. Overall, the structure will be hierarchical.
In such model, it is entirely reasonable to treat a collection of ‘triples’ together, if they form a logical whole that is semantically useful. For example, we might have a first-class attribute that could be used in a clinical encounter to record “family history”. As a first-class property, it could also be used in other contexts, but we might expect that family history property to itself be made up of a collection of other properties, including a list of diagnoses.
Simplified, this might look like: (each entity is a tuple (namespace/value))
Encounter -> Has Family History -» [ Diabetes mellitus, Hypertension, Ischaemic Heart Disease]
If you are familiar with SNOMED CT, you will recognise these semantics are similar to the normalised form of a concept. SNOMED CT has concepts which can themselves be decomposed into a normal form because they represent a complex concept, such as “Family history of diabetes”. It is not difficult to imagine that we can decompose and normalize arbitrary SNOMED concepts and indeed more complex expressions into the wider information model and even round-trip between decomposed and composed forms in order to arbitrarily fatten or flatten the tree.
Such an approach depends very much on having a set of sane clinically meaningful and ontologically correct relationships (attributes) defined for concepts, and an understanding that different attributes might provide a different “view” of the same information.
Computable and declarative properties
Using first-class namespaced properties permits making health and care data more easily computable, because one might have multiple properties that use the same source data to derive a semantically different value. It permits create a facade of well-known properties in front of legacy data, or data in different formats.
As a result, we can define properties to be computed, binding the resolution of such properties at runtime and enabling powerful runtime polymorphism; this means our software can potentially. become capable of processing new types of information without needing to update the code.
For example, simple application might only want to examine a flat list of family history diagnoses and problems, while a more sophisticated application might want to drill-down into other attributes to understand, for example, whether there is a maternal history of a specific condition or not, if that data even existed. It means different namespace/value (ie attribute) resolvers can provide a different ‘view’ of the same underlying data. We move from an approach based on create/read/update/delete (CRUD) semantics to one that separates the reads and writes. We have different models for reads compared to writes, because they are semantically different.
For example, we might represent a current list of medications on the ward as a simple collection, containing entries each of which represents a medication. In each, there would be an encoding of the medication using a UK dm+d identifier as well as dosing information. We might write a resolver that provides a computed property that derives total daily dose from that entry to allow easy graphing of daily doses over time.
Current primary care systems use product-based prescribing and a textual description of dosing. Hospital based systems generally use dose-based prescribing. It is possible to map between these two systems, and so we might also permit resolution of a dose property for product-based prescriptions where possible. Such code can simply operate on pure data structures, is easily testable for safety, can work in isolation and demonstrates that we can add additional functionality to existing systems, and existing data, without needing to change the original.
In this way, health and care software could become entirely decoupled, with pure functions provided by external entities processing our semantically-encoded health and care information: testable, modular, decoupled and deterministic.
Likewise, we can see Address classes in openEHR defined for an organisation, with attributes representing properties not applicable to all countries such as a “Delivery point indicator”, which apparently is something important in Brazil. Because such attributes are not first-class, one ends up with multiple classes representing pretty much the same thing. You can easily end up with an explosion of classes, often in a rigid hierarchy.
Instead, certainly for the UK, it would be possible to register, and then resolve, for example, an LSOA identifier into and then from a variety of contexts including organisations and patients, by runtime polymorphism and runtime registration of resolvers of types, while not impacting on the use of other address attributes in other countries in which LSOA has no meaning.
For instance, we might try to resolve on a patient using a first-class name for that attribute such as “http://ons.gov.uk/Id/LSOA” and know that, given a date, we can use the address history and walk our graph to get the postcode and resolve the LSOA from there. We can have original data distributions that provide these data, such as the UK’s NHS postal code directory, and make use of those data to provide dynamic LSOA properties for the address history of an individual patient, and then link to indices of multiple deprivation, to resolve changes in deprivation over time or after a diagnosis.
First-class attributes means we focus on semantics. We should be able to resolve the value of a property such as http://www.w3.org/2004/02/skos/core#prefLabel and obtain the preferred label for that structure, such as an organisation, or person, or even diagnosis, without necessarily knowing about whether the source information is provided in a proprietary format, HL7 v2, HL7 FHIR, openEHR, UK NHS ODS or a social care system. We prioritise meaning over classes and brittle inheritance patterns that are usually dependent on a specific system or programming language.
Similarly, different data collection “forms” can give the same semantic results, such as the “EDSS” scale for multiple sclerosis can be directly recorded or be part of an extended EDSS-FS scale. Instead of using static class hierarchies, we can define the same semantic property for the overall result and later support other scales that can provide the same semantic result, for example, when drawing a graph of results over time, or working out disease trajectory.
You might recognise the approach of an application walking a graph and requesting exactly the information it requires, as that of GraphQL or Pathom. Clients see the view of data that they request.
You might also recognise that approach of separating the semantics of reading (and making sense of the record) from the writing of information as that of CQRS (Command Query Responsibility Segregation). Managing medication is a good example, because actually health care data is actually better represented as a transactional log of events. Do we really change that dose of medication in a database table or are the semantics better represented as a model of that change instead? Isn’t our current state simply a snapshot of all of the events processed sequentially?
The names and values of data are orthogonal to the specifications of that data, but most object-orientated specifications conflate the two concepts.
Weeknotes
So during my convalescence, I learnt Clojure and also either started or continued work on the following:
-
Concierge. This resolves identifiers using namespace value tuples to abstract away some of NHS Wales’ existing services. This was written in golang and returns HL7 FHIR like models for the resolved resources, providing mapping between different namespaces (e.g. Read codes to SNOMED CT). This work made me realise that first-class types were not as scalable as I’d like, and it would be difficult to provide the same semantics as standards such as HL7 FHIR and its extensions without first-class namespaced and specified attributes.
-
e-Obs. A lightweight clojurescript single page application with desktop application responsiveness in the browser, dynamically generated SVG national early warning score charts on demand, and using concierge to handle user login, security token generation and refresh, and patient lookup. This uses an event-driven architecture. This taught me a lot about functional programming.
-
clods. A clojure microservice providing a RESTful and graph API to UK health and care organisational services and postcodes. This led to me looking at how I might represent proprietary ODS schemas as both FHIR models and the W3C organisation schema, and how it would be so much easier if I used namespaced attributes instead.
-
go-terminology. A very fast golang-based terminology server that provides SNOMED CT services via REST and gRPC APIs. SNOMED-CT is an ontology, and uses triples in the same way as RDF, but the learning here is the importance of mapping concrete values from one ontology to another.
I plan to federate information from ‘clods’ on hospital sites with that from individual patient administration systems to allow hopping from arbitrary sites down into individuals units, wards and individual beds. For the client application, they will simply walk a graph from an organisation to a site, to a unit, to a ward and to a bed, with first-class namespaced attributes providing an abstraction over the inherent complexity in providing access to these data, which are obviously sourced from multiple disparate systems each using different data structures and representations.
I’ve started work on a rule-based declarative system to resolve identifiers arbitrarily in the context of different information models, including making use of the existing and open-source openEHR archetype definitions, and linking those to HL7 FHIR and v2 messages when possible. Mostly those resolvers exist within the same application, at the moment, but there would be no different if those resolvers were provided from an external API, operating on pure data, in order to resolve the identifier in the context provided.
We need to reboot the work on linked data and ontologies and use that in health and care. All of this work should be open-source, because semantically-meaningful data and ontologies are the way we should be building health and care software fit for the 21st Century.
Mark