Practical considerations for using SNOMED CT for analytics and machine learning
Update 2021: You can find the latest version of my SNOMED CT library and microservice: hermes
Introduction
Data-driven healthcare means using data to support and provide health services.
Personal medical data includes current and past medical problems, past and present treatments and interventions but we also increasingly recognise the value in data derived from personal devices and home monitoring. In addition, we need to take into account more generic medical information such as appropriate guidelines, literature and knowledge support.
We need to support both patients and professionals in finding and making sense of that information in order to support decision-making. Similarly, we need to use these data to care not only for an individual patient but to look across groups of patients to understand outcomes and the effects of our interventions at a population-level.
SNOMED CT is a comprehensive healthcare terminology; the UK distribution of SNOMED CT contains over 1,500,000 descriptions of over individual 500,000 concepts comprising problems, symptoms, signs, diagnoses, medications (dm+d) and procedures as well as supporting administrative concepts such as occupations, countries of birth and many others. SNOMED CT has been derived, in part, from the UK Read codes, widely used in general practice, but importantly, while Read codes provide only a single hierarchy, SNOMED CT supports the definition of multiple types of relationship between concepts. The most frequently used relationship, IS-A, provides a taxonomic hierarchy in which specific concepts, such as “myocardial infarction” are classified as a member of a higher class, such as “ischaemic heart disease” or “disorder of the heart”. As a result, SNOMED CT provides a medical ontology that is meaningful to professionals and is computable; I have previously demonstrated my own electronic patient record software that, using code, can systematically identify patients with a certain disease and highlight important outcome measures and compare disease progression with peers in conditions like motor neurone disease and multiple sclerosis (see “so I built an EPR”).
While its size means that it is often very comprehensive, SNOMED CT can be difficult to implement technically and much effort is required to manage the quality of concepts and the relationships between them.
Traditionally, one of the ways of managing the complexity has been to set up a working group of interested clinicians in order to define a reference set of curated terms in order to simplify the implementation for specific clinical workflows. Indeed, there are now over 250 reference sets in the UK distribution alone. Some of these, such as language reference sets, are now an essential part of the SNOMED CT model as they provide the data to support logic on which preferred synonym to use for a particular language or dialect. However, some simply provide a restricted list of concepts to aid implementation, user interface design and analysis for a specific clinical context or subspecialty. For instance, we now have a UK emergency care diagnosis reference set containing 770 clinical diagnoses that are thought to be useful in an emergency department.
I want to argue that,
- we should be spending our time as a community improving the quality of the concepts and relationships in the core SNOMED CT model in preference to thinking we should start by creating a new reference set that will need ongoing maintenance and curation; why unnecessarily limit choice? We can permit access to end-users the whole of SNOMED CT by making use of the taxonomic hierarchy to limit choice in a specific context as long as you design an appropriate user interface; we can limit choice to types of “countries of birth” for a country of birth data entry field and “diagnoses” for a diagnosis data entry field but why constrain only to a subset of diagnoses? The only use of optional constraints is to make it easier for the user to enter commonly used terms, and there are probably better ways of learning those that starting a new committee, such as tracking terms automatically and learning on-the-fly.
- we can support analytics by using that taxonomic hierarchy to simplify and make sense of raw data; in essence, to reduce complex high-dimensionality data into something simpler and computable.
My final point is:
- we need to build open-source tools to build an infrastructure that can act as a platform onto which more innovation, including the application of machine learning, can be built. We need to make this core functionality a commodity.
Using SNOMED CT in real-life
I embedded SNOMED-CT into an electronic patient record system in 2009 and have used it since as the lingua-franca in handling semantic interoperability and appropriate isolation between modules of the application. An article briefly describing its functionality is available here.
There are three important learning points:
- it is possible to implement SNOMED CT and make it accessible to users
- in my experience, you know you have good user interface design when your users do not actually know that they are using SNOMED CT
- the use of SNOMED CT does not mean you enter terms only by direct user entry such as an autocomplete textfield or drop-down selections. SNOMED terms can be imputed as a result of other processes and as such, it can be used as an intermediary structure onto which to build clinical decision support.
Here is a user entering a country of birth:
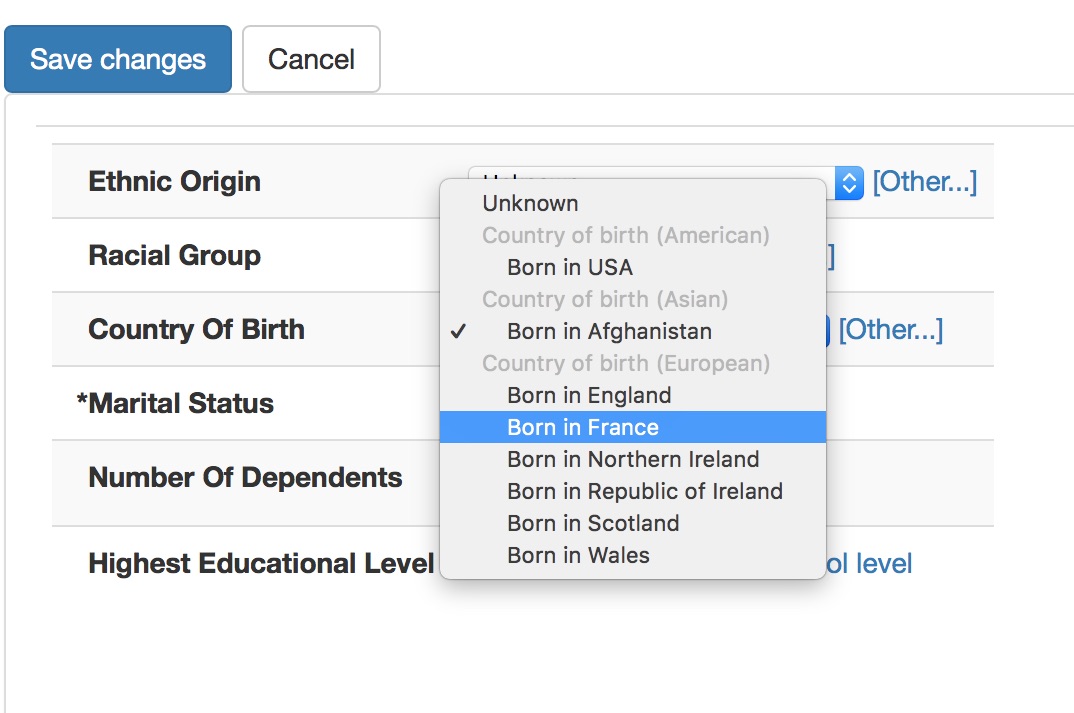
In this diagram, the user has no idea that they are entering SNOMED CT concepts, but their choice is constrained appropriately so that they cannot enter a diagnosis in a country of birth field and we can understand that being born in France means that a patient was born in Europe. I’ve obviously not hard-coded this logic, I’ve simply configured my user interface control to allow:
- selection of a commonly selected concept in this specific context, such as a particular type of clinical setting
- an ability to search for less frequently used concepts with a simple autocomplete search - the “Other…” link opens this link when the user wishes to select a term not in the most-frequently used pop-up.
- constraint of both to the appropriate context, in this case, countries of birth
Here I type MI and can readily select the diagnosis I wanted, in this case “myocardial infarction”. This is from a simple demo application rather than a proper electronic health record:
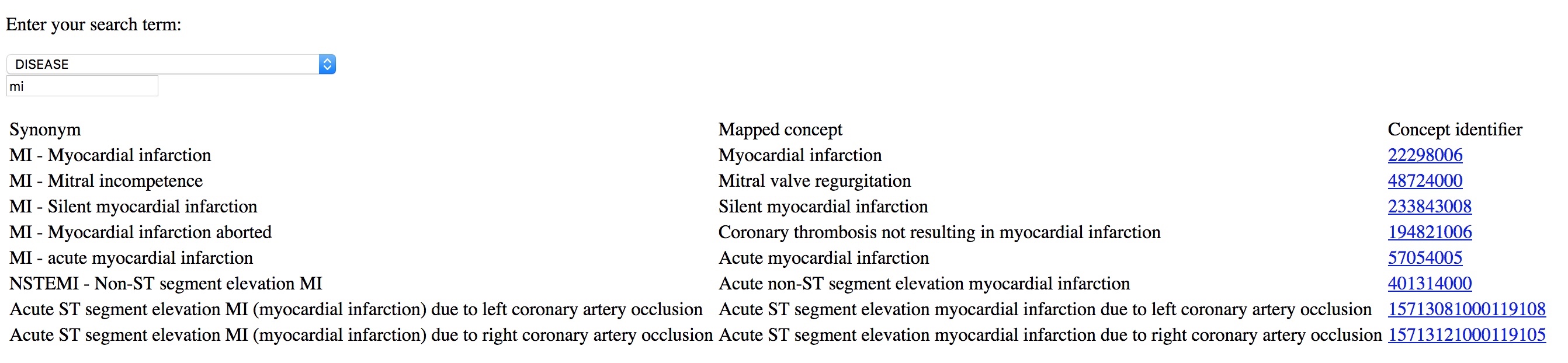
I use the same approach to simplify building clinical forms for data entry, with SNOMED CT terms either entered by the user:
or imputed based on logic from other data entered, such as “able to walk” based on entry of a disease-specific structured disability rating scale. Other modules do not need to ‘understand’ the disease-specific scale but use the intermediary SNOMED based data structures to make appropriate inferences.
If you give tools to professionals that make it easier to enter coded structured information than free-text, they will do so. Coding diagnoses, procedures and background information is critical in understanding outcome data; they are a foundation on which more complex and sophisticated analytics can be built, particularly if you ensure that those analytics are immediately available to support clinical decision making.
Analytics
What is the point of collecting data if we don’t use them to support our clinical care? We must make data that is computable, that is, to be processed and analysed so it is useful for decision making.
Computer-intensive analytics, just like more conventional statistic approaches, can struggle with high-dimensional data. Much of our work in data analytics is cleaning data in order to make it computable. In conventional linear modelling for example, we might hypothesise that a range of covariates, a range of factors, might influence an outcome measurement and so we might estimate that we will need a sample size of 3-20x the number of variables. However, determination of sample size is much more complex than that, needing to take into account effect size and degrees of freedom for each covariate in our regression.
What are our options here?
- Let me enter free-text
- Ask me to record, manually, a general categorical data-entry instead of a specific concept.
- Let me choose from a limited number of “choice” options.
- Record the best, most specific concept at the time of data-entry that matches what I’m thinking
For example, we might have a referral pathway that uses free-text which makes it possible for human consumption when processing individual referrals but difficult to compute, aggregate or make sense when examining many thousands of referrals to understand demands on our services.
Instead, we could ask the referring professional to simply choose a type of referral, such as “ischaemic heart disease” or “valvular heart disease” so that the referral can be redirected to the appropriate subspecialty cardiac service. This limits what we collect so that it becomes easier to process, both for human consumption and for analytic purposes.
There are problems with this approach:
- Categorisation needs to be done prospectively.
- We deliberately lose specific information in favour of recording a general category only.
- It adds an additional step for users.
- If we change our categorisation because of changes in our service, we have to change what the users have to choose and subsequently deal with handling analytics before and after the change.
I believe the best approach is to preserve the granularity, the specificity, of the clinical concept(s) chosen at source and permit arbitrary categorisation at a later date, depending on need.
Open source tooling
I have released two open-source SNOMED CT terminology servers which support both the fast free-text search of SNOMED CT concepts as well as support for analytics and generalisation of specific SNOMED concepts into more general broad diagnoses or categories:
- rsterminology has its roots in 2007-2008 and used to be a module within a larger application; it is now a java microservice built using Apache Cayenne, Bootique and LinkRest. It supports SNOMED distributions in the RF1 release format, which is now deprecated.
- go-terminology is a new golang command-line tool and microservice whose main focus is in supporting analytics. It supports SNOMED RF2, the current release format. It is a work-in-progress but usable now. It is my first golang application.
Obtaining simple information about a concept
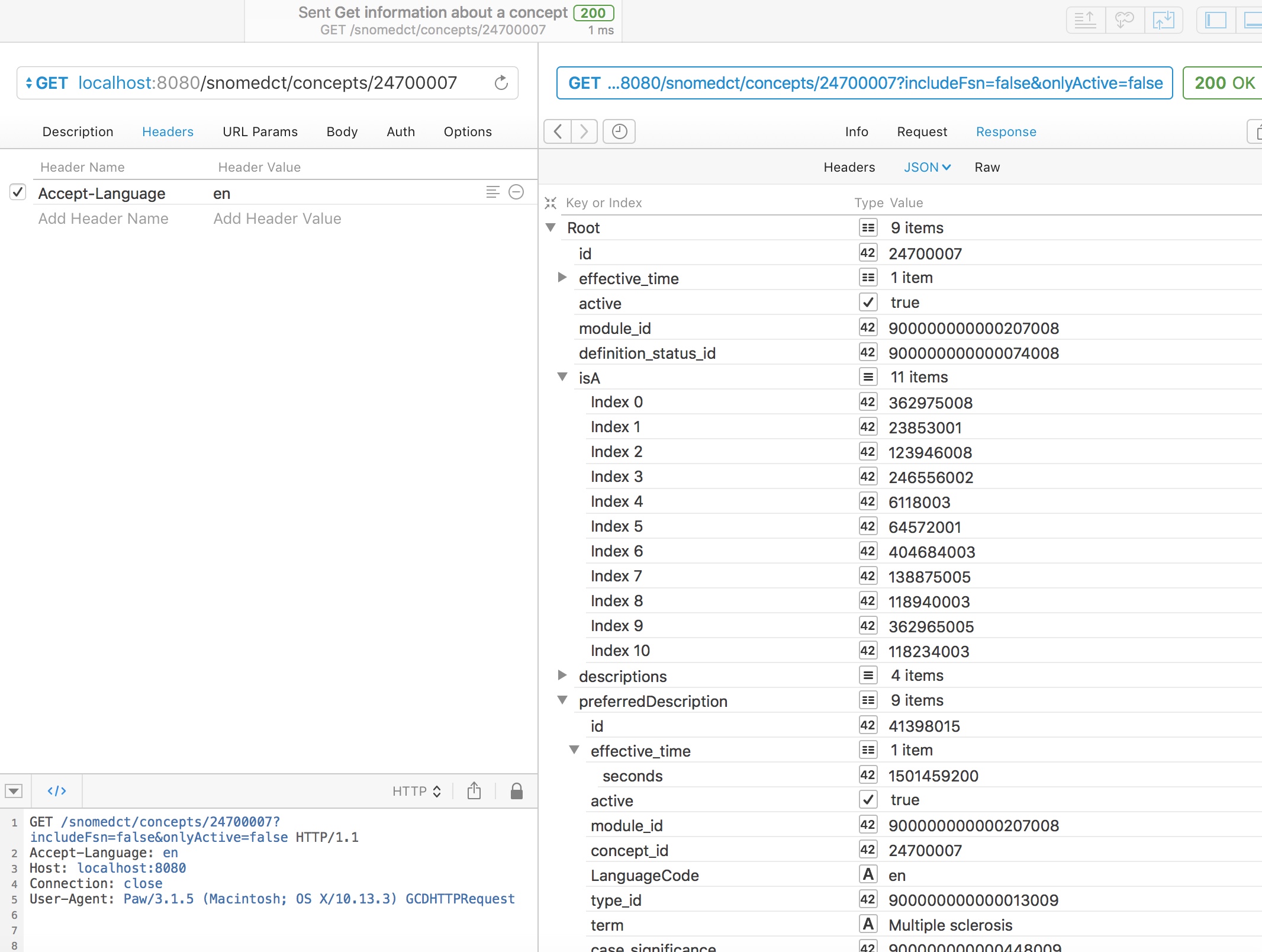
Using either web service, we can readily obtain information about a concept, including the preferred synonym for a particular locale, here British English, and simplified subsumption testing, showing all of the IS-A relationships for this concept walking all of the possible paths up to root.
In this example, we see that multiple sclerosis (24700007) is a type of demyelinating central nervous system disease (6118003) and a type of neurological disease (118940003)
Genericise a concept
Genericising a concept finds the best equivalent match for a specified concept either from a manually chosen list of categories or from an existing refset. This requires the new go-terminology service.
For example, here we take a chosen concept, multiple sclerosis, and map it to the curated UK emergency care diagnosis reference set (991411000000109):
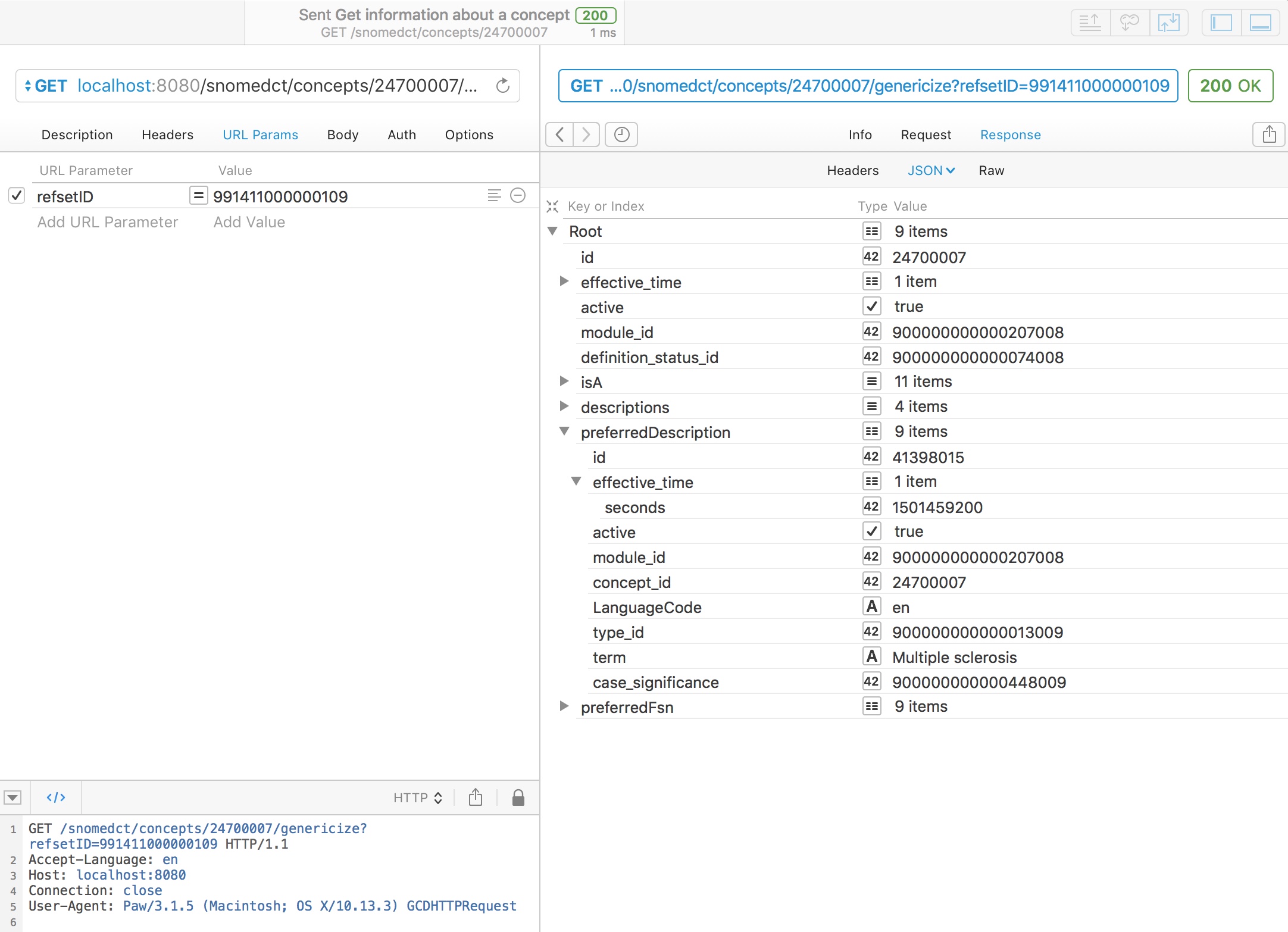
As you might expect, multiple sclerosis is in that refset, so the service simply returns it.
However, a rare disorder like acute disseminated encephalomyelitis (ADEM - 83942000) is not a member of the UK emergency diagnosis reference set. Can we genericise this to make it useful for our analytics? Indeed we can, and using the SNOMED hierarchy, we can determine that ADEM is a type of encephalitis and automatically return that.
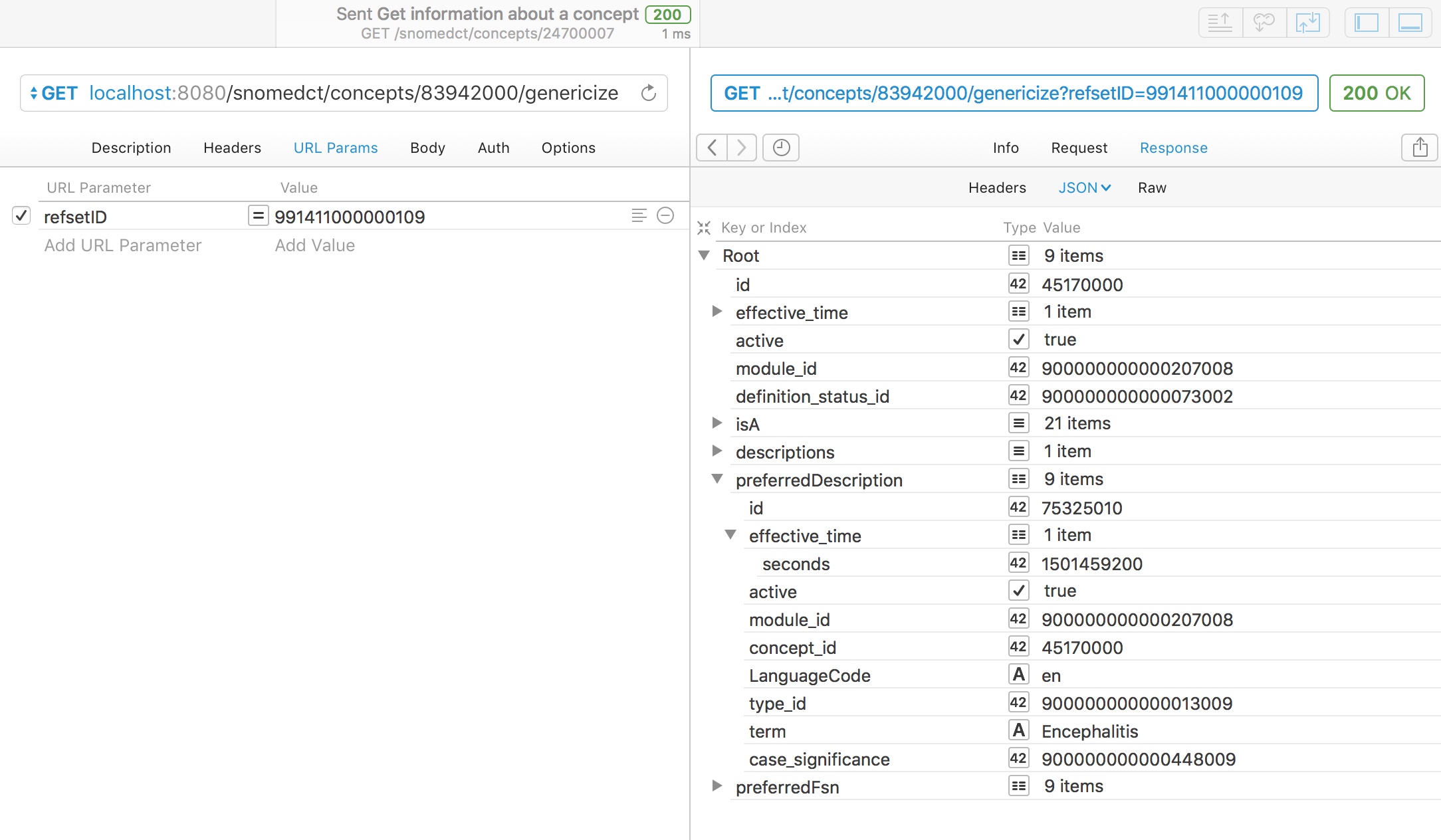
It is important to note that adequate genericisation needs to identify the best match and not just any match as otherwise, ADEM might simply be classified as a neurological disorder; while true, a more specific match exists in that reference set.
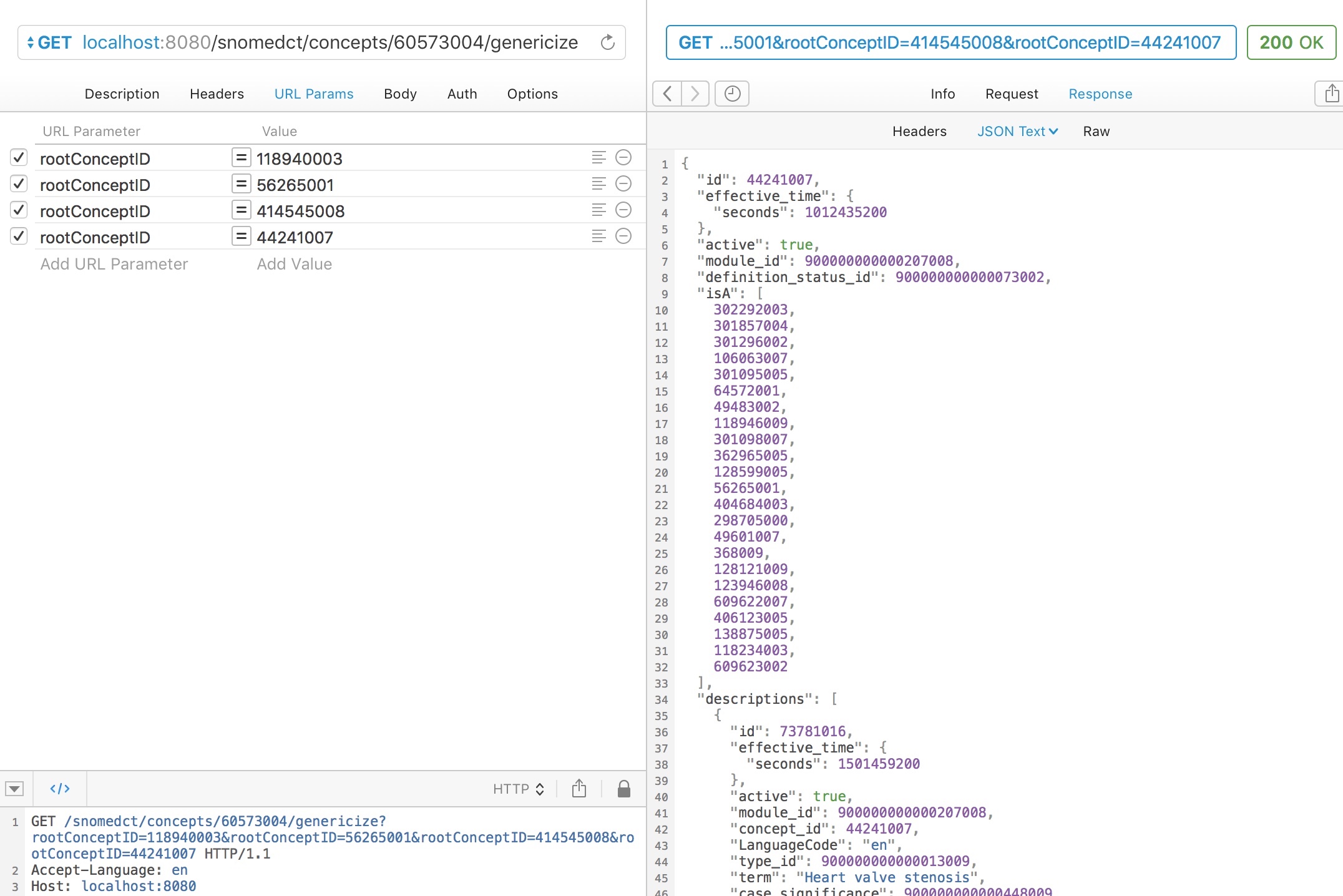
For example, here we ask to genericise “myocardial infarction” (22298006) to one of four broad categories:
- Neurological disorder (118940003)
- Heart disease (56265001)
- Ischaemic heart disease (414545008)
- Heart valve stenosis (44241007)
A simple subsumption test would indicate that myocardial infarction could be either (2) or (3), so what result do we get?
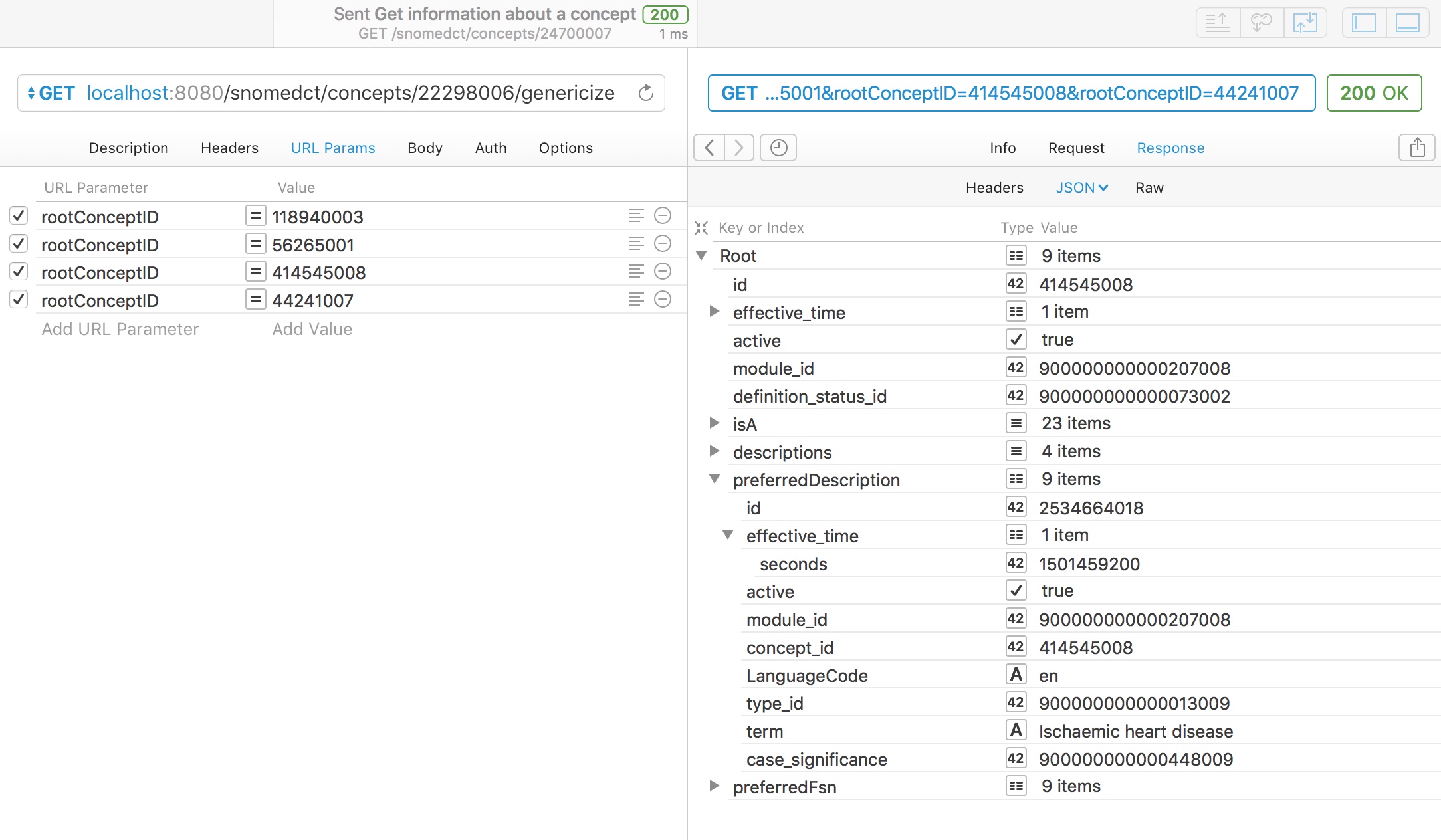
Appropriately, the service maps to “ischaemic heart disease” as the closest best match of the four categories available as it is more specific than the more general term “heart disease”, which subsumes it.
Similarly, aortic stenosis is appropriately mapped to a type of heart valve stenosis.
Conclusions
I have successfully deployed SNOMED CT in real-life clinical environments since 2009, with almost all users entirely unaware that they were using SNOMED CT. I would argue that it is possible to safely permit access to all concepts with limits only on the basis of the taxonomic hierarchy and not on the basis of reference sets. I do limit the initial pop-up list to concepts most frequently used in a specific clinical context and might consider using an existing reference set as a starting list for those. While reference sets are now vital for some functionality, such as supporting localisation, I am sometimes disapppointed by what appears to be the default initial thoughts of working groups and committees, on starting to consider SNOMED CT implementation, to think about defining a new reference set for their purposes. Instead, we should improve the quality of the overall product.
For decision support, analytics and benchmarking our services to others, we can use the taxonomic hierarchy to “understand” the specific concepts entered by our users, mapping them to higher level, broad concepts; we need simple tools to process and simplify structured clinical information, made available to multiple data processing pipelines.
We need to create an open-source foundation of services with which to build the next generation of advanced and valuable clinical information systems. We need to make an open and free infrastructure on which we create value, adopting open-source and building communities in order to create a commodity platform.
To this end, I hope to continue to build these open-source tools to support complex data transformation and simplification and would be keen to get collaborators who wish to do the same. Much of the go-terminology tool is to support the work I outlined in my diagnostic inference blog post), particularly the need for dimensionality reduction in machine learning. Let me know if you want to help.
Mark